Integrated analysis of global mRNA and protein expression data in HEK293 cells overexpressing PRL-1
- PMID: 24019887
- PMCID: PMC3760866
- DOI: 10.1371/journal.pone.0072977
Integrated analysis of global mRNA and protein expression data in HEK293 cells overexpressing PRL-1
Abstract
Background: The protein tyrosine phosphatase PRL-1 represents a putative oncogene with wide-ranging cellular effects. Overexpression of PRL-1 can promote cell proliferation, survival, migration, invasion, and metastasis, but the underlying mechanisms by which it influences these processes remain poorly understood.
Methodology: To increase our comprehension of PRL-1 mediated signaling events, we employed transcriptional profiling (DNA microarray) and proteomics (mass spectrometry) to perform a thorough characterization of the global molecular changes in gene expression that occur in response to stable PRL-1 overexpression in a relevant model system (HEK293).
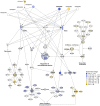
Principal findings: Overexpression of PRL-1 led to several significant changes in the mRNA and protein expression profiles of HEK293 cells. The differentially expressed gene set was highly enriched in genes involved in cytoskeletal remodeling, integrin-mediated cell-matrix adhesion, and RNA recognition and splicing. In particular, members of the Rho signaling pathway and molecules that converge on this pathway were heavily influenced by PRL-1 overexpression, supporting observations from previous studies that link PRL-1 to the Rho GTPase signaling network. In addition, several genes not previously associated with PRL-1 were found to be significantly altered by its expression. Most notable among these were Filamin A, RhoGDIα, SPARC, hnRNPH2, and PRDX2.
Conclusions and significance: This systems-level approach sheds new light on the molecular networks underlying PRL-1 action and presents several novel directions for future, hypothesis-based studies.
Conflict of interest statement
Figures


Similar articles
-
Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer.Br J Cancer. 2006 Aug 7;95(3):347-54. doi: 10.1038/sj.bjc.6603261. Epub 2006 Jul 11. Br J Cancer. 2006. PMID: 16832410 Free PMC article.
-
Expression of phosphatase of regenerating liver 1 and 3 mRNA in esophageal squamous cell carcinoma.Arch Pathol Lab Med. 2008 Aug;132(8):1307-12. doi: 10.5858/2008-132-1307-EOPORL. Arch Pathol Lab Med. 2008. PMID: 18684031
-
PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility.Cancer Res. 2006 Mar 15;66(6):3153-61. doi: 10.1158/0008-5472.CAN-05-3116. Cancer Res. 2006. PMID: 16540666
-
PRL phosphatases as potential molecular targets in cancer.Mol Cancer Ther. 2005 Nov;4(11):1653-61. doi: 10.1158/1535-7163.MCT-05-0248. Mol Cancer Ther. 2005. PMID: 16275986 Review.
-
Molecular mechanisms of the PRL phosphatases.FEBS J. 2013 Jan;280(2):505-24. doi: 10.1111/j.1742-4658.2012.08565.x. Epub 2012 Apr 10. FEBS J. 2013. PMID: 22413991 Review.
Cited by
-
Integrated proteomics and transcriptomics analyses identify novel cell surface markers of HIV latency.Virology. 2022 Aug;573:50-58. doi: 10.1016/j.virol.2022.06.003. Epub 2022 Jun 4. Virology. 2022. PMID: 35714458 Free PMC article.
-
EIF6 over-expression increases the motility and invasiveness of cancer cells by modulating the expression of a critical subset of membrane-bound proteins.BMC Cancer. 2015 Mar 15;15:131. doi: 10.1186/s12885-015-1106-3. BMC Cancer. 2015. PMID: 25886394 Free PMC article.
-
Bottom-up, integrated -omics analysis identifies broadly dosage-sensitive genes in breast cancer samples from TCGA.PLoS One. 2019 Jan 17;14(1):e0210910. doi: 10.1371/journal.pone.0210910. eCollection 2019. PLoS One. 2019. PMID: 30653567 Free PMC article.
-
Identifying three-dimensional structures of autophosphorylation complexes in crystals of protein kinases.Sci Signal. 2015 Dec 1;8(405):rs13. doi: 10.1126/scisignal.aaa6711. Sci Signal. 2015. PMID: 26628682 Free PMC article.
References
-
- Diamond RH, Peters C, Jung SP, Greenbaum LE, Haber BA, et al. (1996) Expression of PRL-1 nuclear PTPase is associated with proliferation in liver but with differentiation in intestine. Am J Physiol 271: G121–129. - PubMed
-
- Guo K, Li J, Wang H, Osato M, Tang JP, et al. (2006) PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Res 66: 9625–9635. - PubMed
-
- Guo S, Russo IH, Russo J (2003) Difference in gene expression profile in breast epithelial cells from women with different reproductive history. Int J Oncol 23: 933–941. - PubMed
-
- Takano S, Fukuyama H, Fukumoto M, Kimura J, Xue JH, et al. (1996) PRL-1, a protein tyrosine phosphatase, is expressed in neurons and oligodendrocytes in the brain and induced in the cerebral cortex following transient forebrain ischemia. Brain Res Mol Brain Res 40: 105–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

