Basic amino acid mutations in the nuclear localization signal of hibiscus chlorotic ringspot virus p23 inhibit virus long distance movement
- PMID: 24019944
- PMCID: PMC3760818
- DOI: 10.1371/journal.pone.0074000
Basic amino acid mutations in the nuclear localization signal of hibiscus chlorotic ringspot virus p23 inhibit virus long distance movement
Abstract
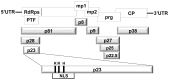
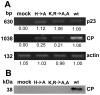
The p23 is a unique protein in the Hibiscus chlorotic ringspot virus which belongs to Family Tombusviridae Genus Carmovirus. Our previous results showed that the p23 is indispensable for host-specific replication and is localized in the nucleus with a novel nuclear localization signal. To investigate additional function(s) of p23, mutations of basic amino acids lysine (K), arginine (R) and histidine (H) that abolish its nuclear localization, were introduced into a biologically active full-length cDNA clone p223 of HCRSV for testing its effects on virus replication and virus movement in vivo. Primer-specific reverse transcription-PCR was conducted to detect gene transcript level of p23 and viral coat protein separately. Virus replication and its coat protein expression were detected by fluorescent in situ hybridization and Western blot, respectively. The effect of p23 was further confirmed by using artificial microRNA inoculation-mediated silencing. Results showed that the two mutants were able to replicate in protoplasts but unable to move from inoculated leaves to newly emerged leaves. Both the p23 and the CP genes of HCRSV were detected in the newly emerged leaves of infected plants but CP was not detected by Western blot and no symptom was observed on those leaves at 19 days post inoculation. This study demonstrates that when p23 is prevented from entering the nucleus, it results in restriction of virus long distance movement which in turn abrogates symptom expression in the newly emerged leaves. We conclude that the p23 protein of HCRSV is required for virus long distance movement.
Conflict of interest statement
Figures


References
-
- You XJ, Kim JW, Stuart GW, Bozarth RF (1995) The nucleotide sequence of Cowpea mottle virus and its assignment to the genus Carmovirus. J Gen Virol 76 2841–2845. - PubMed
-
- Riviere CJ, Rochon DM (1990) Nucleotide sequence and genomic organization of Melon necrotic spot virus. J Gen Virol 71 1887–1896. - PubMed
-
- Berthome R, Kusiak C, Renou JP, Albouy J, Freire MA, et al. (1998) Relationship of the Pelargonium flower break carmovirus (PFBV) coat protein gene with that of other carmoviruses . Arch Virol 143: 1823–1829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

