Unraveling the evolution of the Atlantic cod's (Gadus morhua L.) alternative immune strategy
- PMID: 24019946
- PMCID: PMC3760826
- DOI: 10.1371/journal.pone.0074004
Unraveling the evolution of the Atlantic cod's (Gadus morhua L.) alternative immune strategy
Erratum in
- PLoS One. 2013;8(10). doi:10.1371/annotation/18b70612-fd3d-46ce-a04b-652d18c82d5b
Abstract
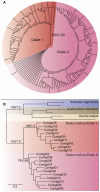
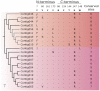
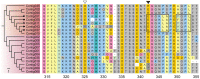
Genes encoding the major histocompatibility complex (MHC) have been thought to play a vital role in the adaptive immune system in all vertebrates. The discovery that Atlantic cod (Gadus morhua) has lost important components of the MHC II pathway, accompanied by an unusually high number of MHC I genes, shed new light on the evolution and plasticity of the immune system of teleosts as well as in higher vertebrates. The overall aim of this study was to further investigate the highly expanded repertoire of MHC I genes using a cDNA approach to obtain sequence information of both the binding domains and the sorting signaling potential in the cytoplasmic tail. Here we report a novel combination of two endosomal sorting motifs, one tyrosine-based associated with exogenous peptide presentation by cross-presenting MHCI molecules, and one dileucine-based associated with normal MHC II functionality. The two signal motifs were identified in the cytoplasmic tail in a subset of the genes. This indicates that these genes have evolved MHC II-like functionality, allowing a more versatile use of MHC I through cross-presentation. Such an alternative immune strategy may have arisen through adaptive radiation and acquisition of new gene function as a response to changes in the habitat of its ancestral lineage.
Conflict of interest statement
Figures


Similar articles
-
Antigen presentation genes in gadoid species (haddock: Melanogrammus aeglefinus and Atlantic cod: Gadus morhua) raise questions about cross-presentation pathways and glycosylated beta-2-microglobulin.Mol Immunol. 2021 Jan;129:21-31. doi: 10.1016/j.molimm.2020.11.011. Epub 2020 Nov 28. Mol Immunol. 2021. PMID: 33260037
-
Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats.BMC Genomics. 2018 Apr 10;19(1):240. doi: 10.1186/s12864-018-4616-y. BMC Genomics. 2018. PMID: 29636006 Free PMC article.
-
Characterization of MHC class I and beta(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes.Immunogenetics. 1999 Oct;50(1-2):49-59. doi: 10.1007/s002510050685. Immunogenetics. 1999. PMID: 10541806
-
Novel molecules related to MHC antigens.Curr Opin Immunol. 1995 Feb;7(1):97-102. doi: 10.1016/0952-7915(95)80034-4. Curr Opin Immunol. 1995. PMID: 7772287 Review.
-
Evolutionary conservation of MHC class I and class II molecules--different yet the same.Semin Immunol. 1994 Dec;6(6):411-24. doi: 10.1006/smim.1994.1050. Semin Immunol. 1994. PMID: 7654997 Review.
Cited by
-
The evolution of the major histocompatibility complex in upstream versus downstream river populations of the longnose dace.Ecol Evol. 2017 Apr 1;7(10):3297-3311. doi: 10.1002/ece3.2839. eCollection 2017 May. Ecol Evol. 2017. PMID: 28515867 Free PMC article.
-
Whole genome sequencing data and de novo draft assemblies for 66 teleost species.Sci Data. 2017 Jan 17;4:160132. doi: 10.1038/sdata.2016.132. Sci Data. 2017. PMID: 28094797 Free PMC article.
-
Single-Cell Transcriptome Profiling of Immune Cell Repertoire of the Atlantic Cod Which Naturally Lacks the Major Histocompatibility Class II System.Front Immunol. 2020 Oct 9;11:559555. doi: 10.3389/fimmu.2020.559555. eCollection 2020. Front Immunol. 2020. PMID: 33154745 Free PMC article.
-
Present Yourself! By MHC Class I and MHC Class II Molecules.Trends Immunol. 2016 Nov;37(11):724-737. doi: 10.1016/j.it.2016.08.010. Epub 2016 Sep 7. Trends Immunol. 2016. PMID: 27614798 Free PMC article. Review.
-
Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions.Sci Rep. 2016 Apr 29;6:25211. doi: 10.1038/srep25211. Sci Rep. 2016. PMID: 27126702 Free PMC article.
References
-
- Schluter SF, Bernstein RM, Bernstein H, Marchalonis JJ (1999) ‘Big Bang’ emergence of the combinatorial immune system. Dev Comp Immunol 23: 107–111. - PubMed
-
- Landsverk OJ, Bakke O, Gregers TF (2009) MHC II and the endocytic pathway: regulation by invariant chain. Scand J Immunol 70: 184–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

