Experimental pathways towards developing a rotavirus reverse genetics system: synthetic full length rotavirus ssRNAs are neither infectious nor translated in permissive cells
- PMID: 24019962
- PMCID: PMC3760874
- DOI: 10.1371/journal.pone.0074328
Experimental pathways towards developing a rotavirus reverse genetics system: synthetic full length rotavirus ssRNAs are neither infectious nor translated in permissive cells
Abstract
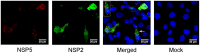
At present the ability to create rationally engineered mutant rotaviruses is limited because of the lack of a tractable helper virus-free reverse genetics system. Using the cell culture adapted bovine RV RF strain (G6P6 [1]), we have attempted to recover infectious RV by co-transfecting in vitro transcribed ssRNAs which are identical in sequence to the positive sense strand of each of the 11 dsRNA genomic segments of the RF strain. The RNAs were produced either from cDNAs cloned by a target sequence-independent procedure, or from purified double layered RV particles (DLPs). We have validated their translational function by in vitro synthesis of (35)S-labelled proteins in rabbit reticulocyte lysates; all 11 proteins encoded by the RV genome were expressed. Transfection experiments with DLP- or cDNA-derived ssRNAs suggested that the RNAs do not act independently as mRNAs for protein synthesis, once delivered into various mammalian cell lines, and exhibit cytotoxicity. Transfected RNAs were not infectious since a viral cytopathic effect was not observed after infection of MA104 cells with lysates from transfected cells. By contrast, an engineered mRNA encoding eGFP was expressed when transfected under identical conditions into the same cell lines. Co-expression of plasmids encoding NSP2 and NSP5 using a fowlpox T7 polymerase recombinant virus revealed viroplasm-like structure formation, but this did not enable the translation of transfected RV ssRNAs. Attempts to recover RV from ssRNAs transcribed intracellularly from transfected cDNAs were also unsuccessful and suggested that these RNAs were also not translated, in contrast to successful translation from a transfected cDNA encoding an eGFP mRNA.
Conflict of interest statement
Figures





References
-
- Estes MK, Kapikian AZ (2007) Rotaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed: Lippincott Williams & Wilkins/Wolters Kluwer, Philadelphia. 1917–1974.
-
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, et al. (2012) 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. The Lancet Infectious Diseases 12: 136–141. - PubMed
-
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, et al. (2006) Safety and Efficacy of a Pentavalent Human-Bovine (WC3) Reassortant Rotavirus Vaccine. New England Journal of Medicine 354: 23–33. - PubMed
-
- Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, et al. (2006) Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. New England Journal of Medicine 354: 11–22. - PubMed
-
- SAGE (2009) Meeting of the immunization strategic advisory group of experts, April 2009 - conclusions and recommendations. Weekly Epidemiological Record 84: 220–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

