A forward genetic approach in Chlamydomonas reinhardtii as a strategy for exploring starch catabolism
- PMID: 24019981
- PMCID: PMC3760859
- DOI: 10.1371/journal.pone.0074763
A forward genetic approach in Chlamydomonas reinhardtii as a strategy for exploring starch catabolism
Abstract
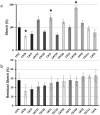
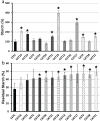
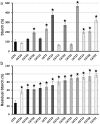
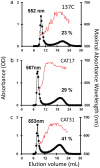
A screen was recently developed to study the mobilization of starch in the unicellular green alga Chlamydomonas reinhardtii. This screen relies on starch synthesis accumulation during nitrogen starvation followed by the supply of nitrogen and the switch to darkness. Hence multiple regulatory networks including those of nutrient starvation, cell cycle control and light to dark transitions are likely to impact the recovery of mutant candidates. In this paper we monitor the specificity of this mutant screen by characterizing the nature of the genes disrupted in the selected mutants. We show that one third of the mutants consisted of strains mutated in genes previously reported to be of paramount importance in starch catabolism such as those encoding β-amylases, the maltose export protein, and branching enzyme I. The other mutants were defective for previously uncharacterized functions some of which are likely to define novel proteins affecting starch mobilization in green algae.
Conflict of interest statement
Figures

References
-
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: : 1126–1149. - PubMed
-
- Fincher GB (1989) Molecular and cellular biology associated with endosperm mobilization in germinating grain cereal grains. Annu Rev Plant Physiol Plant Mol Biol 40: 305–346.
-
- Razem FA, Davis AR (1999) Anatomical and ultrastructural changes of the floral nectary of Pisum sativum L during flower development. Protoplasma 206: 57–72.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

