The diversity of histone versus nonhistone sirtuin substrates
- PMID: 24020006
- PMCID: PMC3764476
- DOI: 10.1177/1947601913483767
The diversity of histone versus nonhistone sirtuin substrates
Abstract
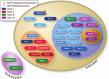
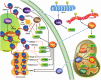
The members of the Sir2 family, or sirtuins, are major regulators of the response to different types of stress. The members of the family have adapted to increasing complexities throughout evolution and have become diversified by increasing their number, specificity, and localization and acquiring novel functions. Sirtuins have been consistently implicated in the cross-talk between the genomic information and environment from the prokaryotes onward. Evidence suggests that in the transition to eukaryotes, histones became one of the basic and most conserved targets of the family, to the extent that in yeast and mammals, sirtuins were originally described as NAD(+)-dependent histone deacetylases and classified as class III histone deacetylases. A growing number of studies have determined that sirtuins also target a wide range of nonhistone proteins. Many of these targets are also directly or indirectly related to chromatin regulation. The number of targets has grown considerably in the last decade but has provoked an ill-founded discussion that neglects the importance of histones as sirtuin targets. In this review, we summarize our knowledge regarding the range of sirtuin targets described to date and discuss the different functional implications of histone and nonhistone targets throughout evolution.
Keywords: SIRT1-SIRT7; deacetylation; epigenetics; genome stability; histones; sirtuins; stress response.
Conflict of interest statement
Figures

References
-
- Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483-8 - PubMed
-
- Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb Symp Quant Biol. 2000;65:297-302 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
