Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human
- PMID: 24021628
- PMCID: PMC3905846
- DOI: 10.1093/nar/gkt770
Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human
Abstract
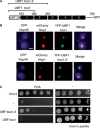
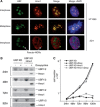
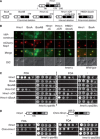
Ribosome biogenesis is a major metabolic effort for growing cells. In Saccharomyces cerevisiae, Hmo1, an abundant high-mobility group box protein (HMGB) binds to the coding region of the RNA polymerase I transcribed ribosomal RNAs genes and the promoters of ∼70% of ribosomal protein genes. In this study, we have demonstrated the functional conservation of eukaryotic HMGB proteins involved in ribosomal DNA (rDNA) transcription. We have shown that when expressed in budding yeast, human UBF1 and a newly identified Sp-Hmo1 (Schizosaccharomyces pombe) localize to the nucleolus and suppress growth defect of the RNA polymerase I mutant rpa49-Δ. Owing to the multiple functions of both proteins, Hmo1 and UBF1 are not fully interchangeable. By deletion and domains swapping in Hmo1, we identified essential domains that stimulate rDNA transcription but are not fully required for stimulation of ribosomal protein genes expression. Hmo1 is organized in four functional domains: a dimerization module, a canonical HMGB motif followed by a conserved domain and a C-terminal nucleolar localization signal. We propose that Hmo1 has acquired species-specific functions and shares with UBF1 and Sp-Hmo1 an ancestral function to stimulate rDNA transcription.
Figures






References
-
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell. Biol. 2009;21:855–863. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

