Optochemical control of RNA interference in mammalian cells
- PMID: 24021631
- PMCID: PMC3905849
- DOI: 10.1093/nar/gkt806
Optochemical control of RNA interference in mammalian cells
Abstract
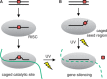
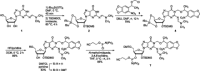
Short interfering RNAs (siRNAs) and microRNAs (miRNAs) have been widely used in mammalian tissue culture and model organisms to selectively silence genes of interest. One limitation of this technology is the lack of precise external control over the gene-silencing event. The use of photocleavable protecting groups installed on nucleobases is a promising strategy to circumvent this limitation, providing high spatial and temporal control over siRNA or miRNA activation. Here, we have designed, synthesized and site-specifically incorporated new photocaged guanosine and uridine RNA phosphoramidites into short RNA duplexes. We demonstrated the applicability of these photocaged siRNAs in the light-regulation of the expression of an exogenous green fluorescent protein reporter gene and an endogenous target gene, the mitosis motor protein, Eg5. Two different approaches were investigated with the caged RNA molecules: the light-regulation of catalytic RNA cleavage by RISC and the light-regulation of seed region recognition. The ability to regulate both functions with light enables the application of this optochemical methodology to a wide range of small regulatory RNA molecules.
Figures





References
-
- Young DD, Deiters A. Photochemical control of biological processes. Org. Biomol. Chem. 2007;5:999–1005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

