A map of cell type-specific auxin responses
- PMID: 24022006
- PMCID: PMC3792342
- DOI: 10.1038/msb.2013.40
A map of cell type-specific auxin responses
Abstract
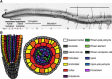
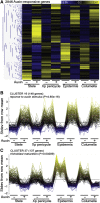
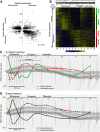
In plants, changes in local auxin concentrations can trigger a range of developmental processes as distinct tissues respond differently to the same auxin stimulus. However, little is known about how auxin is interpreted by individual cell types. We performed a transcriptomic analysis of responses to auxin within four distinct tissues of the Arabidopsis thaliana root and demonstrate that different cell types show competence for discrete responses. The majority of auxin-responsive genes displayed a spatial bias in their induction or repression. The novel data set was used to examine how auxin influences tissue-specific transcriptional regulation of cell-identity markers. Additionally, the data were used in combination with spatial expression maps of the root to plot a transcriptomic auxin-response gradient across the apical and basal meristem. The readout revealed a strong correlation for thousands of genes between the relative response to auxin and expression along the longitudinal axis of the root. This data set and comparative analysis provide a transcriptome-level spatial breakdown of the response to auxin within an organ where this hormone mediates many aspects of development.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
-
- Bennett T, Scheres B (2010) Root development-two meristems for the price of one? Curr Top Dev Biol 91: 67–102 - PubMed
-
- Bhalerao RP, Bennett MJ (2003) The case for morphogens in plants. Nat Cell Biol 5: 939–943 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

