Expanded CD8+ T cells of murine and human CLL are driven into a senescent KLRG1+ effector memory phenotype
- PMID: 24022692
- PMCID: PMC11029347
- DOI: 10.1007/s00262-013-1473-z
Expanded CD8+ T cells of murine and human CLL are driven into a senescent KLRG1+ effector memory phenotype
Abstract
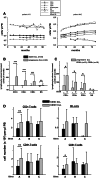
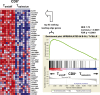
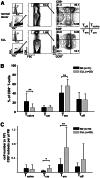
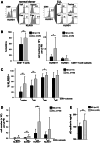
Altered numbers and functions of T cells have previously been demonstrated in chronic lymphocytic leukemia (CLL) patients. However, dynamics and specific T-cell subset alterations have not been studied in great detail. Therefore, we studied CLL blood lymphocyte subsets of individual patients in a longitudinal manner. Dynamic expansions of blood CD4 + and CD8 + T-cell numbers were consistently associated with a progressively increasing CLL leukemic compartment. Interestingly, the T-cell subset expansion over time was more pronounced in CD38 + CLL. Additionally, we performed gene expression profiling of CD3 + T cells of CLL patients and normal donors. Using gene set enrichment analysis, we found significant enrichment of genes with higher expression in CLL T cells within CD8+ effector memory and terminal effector T-cell gene signatures. In agreement with these data, we observed a marked expansion of phenotypic CD8 + effector memory T cells in CLL by flow cytometry. Moreover, we observed that increments of CD8 + effector memory T cells in human CLL and also mouse CLL (Eμ-TCL1 model) were due to an expansion of the inhibitory killer cell lectin-like receptor G1 (KLRG1) expressing cellular subset. Furthermore, higher plasma levels of the natural KLRG1 ligand E-cadherin were detected in CLL patients compared to normal donor controls. The predominance of KLRG1+ expression within CD8+ T cells in conjunction with increased systemic soluble E-cadherin might significantly contribute to CLL immune dysfunction and might additionally represent an important component of the CLL microenvironment.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, Chum P, Yan XJ, Allen SL, Kolitz JE, Baskar S, Rader C, Mellstedt H, Rabbani H, Lee A, Gregersen PK, Rai KR, Chiorazzi N. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood. 2011;117(20):5463–5472. doi: 10.1182/blood-2010-12-324210. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

