Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR
- PMID: 24023039
- PMCID: PMC3831677
- DOI: 10.1002/pro.2368
Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR
Abstract
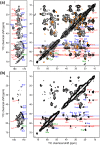
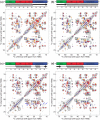
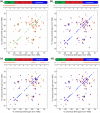
The influenza A M2 protein forms a proton channel for virus infection and mediates virus assembly and budding. While extensive structural information is known about the transmembrane helix and an adjacent amphipathic helix, the conformation of the N-terminal ectodomain and the C-terminal cytoplasmic tail remains largely unknown. Using two-dimensional (2D) magic-angle-spinning solid-state NMR, we have investigated the secondary structure and dynamics of full-length M2 (M2FL) and found them to depend on the membrane composition. In 2D (13)C DARR correlation spectra, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)-bound M2FL exhibits several peaks at β-sheet chemical shifts, which result from water-exposed extramembrane residues. In contrast, M2FL bound to cholesterol-containing membranes gives predominantly α-helical chemical shifts. Two-dimensional J-INADEQUATE spectra and variable-temperature (13)C spectra indicate that DMPC-bound M2FL is highly dynamic while the cholesterol-containing membranes significantly immobilize the protein at physiological temperature. Chemical-shift prediction for various secondary-structure models suggests that the β-strand is located at the N-terminus of the DMPC-bound protein, while the cytoplasmic domain is unstructured. This prediction is confirmed by the 2D DARR spectrum of the ectodomain-truncated M2(21-97), which no longer exhibits β-sheet chemical shifts in the DMPC-bound state. We propose that the M2 conformational change results from the influence of cholesterol, and the increased helicity of M2FL in cholesterol-rich membranes may be relevant for M2 interaction with the matrix protein M1 during virus assembly and budding. The successful determination of the β-strand location suggests that chemical-shift prediction is a promising approach for obtaining structural information of disordered proteins before resonance assignment.
Keywords: chemical shift prediction; conformational change; influenza M2; membrane protein.
© 2013 The Protein Society.
Figures
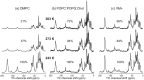



Similar articles
-
Investigation of the curvature induction and membrane localization of the influenza virus M2 protein using static and off-magic-angle spinning solid-state nuclear magnetic resonance of oriented bicelles.Biochemistry. 2015 Apr 7;54(13):2214-26. doi: 10.1021/acs.biochem.5b00127. Epub 2015 Mar 26. Biochemistry. 2015. PMID: 25774685 Free PMC article.
-
Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein.J Am Chem Soc. 2011 Aug 3;133(30):11572-9. doi: 10.1021/ja202051n. Epub 2011 Jul 7. J Am Chem Soc. 2011. PMID: 21661724 Free PMC article.
-
Chemical ligation of the influenza M2 protein for solid-state NMR characterization of the cytoplasmic domain.Protein Sci. 2015 Jul;24(7):1087-99. doi: 10.1002/pro.2690. Epub 2015 May 27. Protein Sci. 2015. PMID: 25966817 Free PMC article.
-
Flu channel drug resistance: a tale of two sites.Protein Cell. 2010 Mar;1(3):246-58. doi: 10.1007/s13238-010-0025-y. Epub 2010 Feb 23. Protein Cell. 2010. PMID: 21203971 Free PMC article. Review.
-
Drug sensitivity, drug-resistant mutations, and structures of three conductance domains of viral porins.Biochim Biophys Acta. 2011 Feb;1808(2):538-46. doi: 10.1016/j.bbamem.2010.07.015. Epub 2010 Jul 23. Biochim Biophys Acta. 2011. PMID: 20655872 Free PMC article. Review.
Cited by
-
Oligomeric Structure and Three-Dimensional Fold of the HIV gp41 Membrane-Proximal External Region and Transmembrane Domain in Phospholipid Bilayers.J Am Chem Soc. 2018 Jul 5;140(26):8246-8259. doi: 10.1021/jacs.8b04010. Epub 2018 Jun 22. J Am Chem Soc. 2018. PMID: 29888593 Free PMC article.
-
Challenges and approaches to understand cholesterol-binding impact on membrane protein function: an NMR view.Cell Mol Life Sci. 2018 Jun;75(12):2137-2151. doi: 10.1007/s00018-018-2789-9. Epub 2018 Mar 8. Cell Mol Life Sci. 2018. PMID: 29520423 Free PMC article. Review.
-
Virus Structures and Dynamics by Magic-Angle Spinning NMR.Annu Rev Virol. 2021 Sep 29;8(1):219-237. doi: 10.1146/annurev-virology-011921-064653. Annu Rev Virol. 2021. PMID: 34586870 Free PMC article. Review.
-
Structural Transition from Closed to Open for the Influenza A M2 Proton Channel as Observed by Proton-Detected Solid-State NMR.J Am Chem Soc. 2025 Aug 6;147(31):27537-27551. doi: 10.1021/jacs.5c05111. Epub 2025 Jun 20. J Am Chem Soc. 2025. PMID: 40539990 Free PMC article.
-
The Influenza M2 Ectodomain Regulates the Conformational Equilibria of the Transmembrane Proton Channel: Insights from Solid-State Nuclear Magnetic Resonance.Biochemistry. 2016 Sep 27;55(38):5387-97. doi: 10.1021/acs.biochem.6b00727. Epub 2016 Sep 12. Biochemistry. 2016. PMID: 27571210 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

