Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting
- PMID: 24023619
- PMCID: PMC3758299
- DOI: 10.1371/journal.pone.0072457
Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting
Abstract
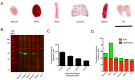
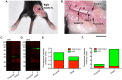
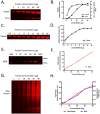
Western blotting has been a key technique for determining the relative expression of proteins within complex biological samples since the first publications in 1979. Recent developments in sensitive fluorescent labels, with truly quantifiable linear ranges and greater limits of detection, have allowed biologists to probe tissue specific pathways and processes with higher resolution than ever before. However, the application of quantitative Western blotting (QWB) to a range of healthy tissues and those from degenerative models has highlighted a problem with significant consequences for quantitative protein analysis: how can researchers conduct comparative expression analyses when many of the commonly used reference proteins (e.g. loading controls) are differentially expressed? Here we demonstrate that common controls, including actin and tubulin, are differentially expressed in tissues from a wide range of animal models of neurodegeneration. We highlight the prevalence of such alterations through examination of published "-omics" data, and demonstrate similar responses in sensitive QWB experiments. For example, QWB analysis of spinal cord from a murine model of Spinal Muscular Atrophy using an Odyssey scanner revealed that beta-actin expression was decreased by 19.3±2% compared to healthy littermate controls. Thus, normalising QWB data to β-actin in these circumstances could result in 'skewing' of all data by ∼20%. We further demonstrate that differential expression of commonly used loading controls was not restricted to the nervous system, but was also detectable across multiple tissues, including bone, fat and internal organs. Moreover, expression of these "control" proteins was not consistent between different portions of the same tissue, highlighting the importance of careful and consistent tissue sampling for QWB experiments. Finally, having illustrated the problem of selecting appropriate single protein loading controls, we demonstrate that normalisation using total protein analysis on samples run in parallel with stains such as Coomassie blue provides a more robust approach.
Conflict of interest statement
Figures

References
-
- Zellner M, Babeluk R, Diestinger M, Pirchegger P, Skeledzic S, et al. (2008) Fluorescence-based Western blotting for quantitation of protein biomarkers in clinical samples. Electrophoresis 29: 3621–3627. - PubMed
-
- Gingrich JC, Davis DR, Nguyen Q (2000) Multiplex detection and quantitation of proteins on western blots using fluorescent probes. Biotechniques 29: 636–42. - PubMed
-
- Suzuki O, Koura M, Noguchi Y, Uchio-Yamada K, Matsuda J (2011) Use of sample mixtures for standard curve creation in quantitative Western blots. Exp Anim 60: 193–196. - PubMed
-
- Wishart TM, Pemberton HN, James SR, McCabe CJ, Gillingwater TH (2008) Modified cell cycle status in a mouse model of altered neuronal vulnerability (slow Wallerian degeneration (Wlds). Genome Biol 9: R101 doi:10.1186/gb-2008-9-6-r101 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

