Evolutionary origin of the Scombridae (tunas and mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families
- PMID: 24023883
- PMCID: PMC3762723
- DOI: 10.1371/journal.pone.0073535
Evolutionary origin of the Scombridae (tunas and mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families
Abstract
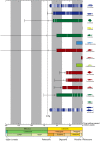
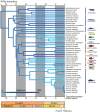
Uncertainties surrounding the evolutionary origin of the epipelagic fish family Scombridae (tunas and mackerels) are symptomatic of the difficulties in resolving suprafamilial relationships within Percomorpha, a hyperdiverse teleost radiation that contains approximately 17,000 species placed in 13 ill-defined orders and 269 families. Here we find that scombrids share a common ancestry with 14 families based on (i) bioinformatic analyses using partial mitochondrial and nuclear gene sequences from all percomorphs deposited in GenBank (10,733 sequences) and (ii) subsequent mitogenomic analysis based on 57 species from those targeted 15 families and 67 outgroup taxa. Morphological heterogeneity among these 15 families is so extraordinary that they have been placed in six different perciform suborders. However, members of the 15 families are either coastal or oceanic pelagic in their ecology with diverse modes of life, suggesting that they represent a previously undetected adaptive radiation in the pelagic realm. Time-calibrated phylogenies imply that scombrids originated from a deep-ocean ancestor and began to radiate after the end-Cretaceous when large predatory epipelagic fishes were selective victims of the Cretaceous-Paleogene mass extinction. We name this clade of open-ocean fishes containing Scombridae "Pelagia" in reference to the common habitat preference that links the 15 families.
Conflict of interest statement
Figures







References
-
- Helfman GS, Collette BB, Facey DE, Bowen BW (2009) The diversity of fishes. Oxford, UK: Wiley-Blackwell. xviii +720 p.
-
- Allen LG, Cross JN (2006) Surface waters. In: Allen LG, Pondella DJ, Horn MH, editors. The ecology of marine fishes: California and adjacent waters. Berkeley: University of California Press. 320–341.
-
- Tyus HM (2012) Ecology and coservation of fishes. London: CRC Press. 529 p.
-
- Froese R, Pauly D (2000) FishBase 2000: concepts, design and data sources. Los Baños, Laguna, Phillipines: ICLARM.
-
- Magnuson JJ (1979) Locomotion by scombrid fishes: hydromechanics, morphology, and behavior. In: Hoar WS, Randall DJ, editors. Fish physiology. London: Academic Press. 239–313.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

