DEAD box protein DDX1 regulates cytoplasmic localization of KSRP
- PMID: 24023901
- PMCID: PMC3762726
- DOI: 10.1371/journal.pone.0073752
DEAD box protein DDX1 regulates cytoplasmic localization of KSRP
Abstract
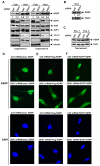
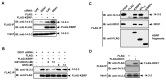
mRNA decay mediated by the AU-rich elements (AREs) is one of the most studied post-transcriptional mechanisms and is modulated by ARE-binding proteins (ARE-BPs). To understand the regulation of K homology splicing regulatory protein (KSRP), a decay-promoting ARE-BP, we purified KSRP protein complexes and identified an RNA helicase, DDX1. We showed that down-regulation of DDX1 expression elevated cytoplasmic levels of KSRP and facilitated ARE-mediated mRNA decay. Association of KSRP with 14-3-3 proteins, that are predominately located in the cytoplasm, increased upon reduction of DDX1. We also demonstrated that KSRP associated with DDX1 or 14-3-3, but not both. These observations indicate that subcellular localization of KSRP is regulated by competing interactions with DDX1 or 14-3-3.
Conflict of interest statement
Figures


References
-
- Wilusz CJ, Wormington M, Peltz SW (2001) The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2: 237-246. doi:10.1038/35067025. PubMed: 11283721. - DOI - PubMed
-
- Wilusz CJ, Wilusz J (2004) Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 20: 491-497. doi:10.1016/j.tig.2004.07.011. PubMed: 15363903. - DOI - PubMed
-
- Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465-470. doi:10.1016/S0968-0004(00)89102-1. PubMed: 8578590. - DOI - PubMed
-
- Bakheet T, Williams BR, Khabar KS (2003) ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res 31: 421-423. doi:10.1093/nar/gkg023. PubMed: 12520039. - DOI - PMC - PubMed
-
- Bakheet T, Williams BR, Khabar KS (2006) ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res 34: D111-D114. doi:10.1093/nar/gkj052. PubMed: 16381826. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

