Molecular and phylogenetic analysis of the porcine kobuvirus VP1 region using infected pigs from Sichuan Province, China
- PMID: 24025093
- PMCID: PMC3847588
- DOI: 10.1186/1743-422X-10-281
Molecular and phylogenetic analysis of the porcine kobuvirus VP1 region using infected pigs from Sichuan Province, China
Abstract
Background: Porcine kobuvirus (PKoV) is a member of the Kobuvirus genus within the Picornaviridae family. PKoV is distributed worldwide with high prevalence in clinically healthy pigs and those with diarrhea.
Methods: Fecal and intestinal samples (n = 163) from pig farms in Sichuan Province, China were obtained to determine the presence of PKoV using reverse transcription polymerase chain reaction assays. Specific primers were used for the amplification of the gene encoding the PKoV VP1 protein sequence. Sequence and phylogenetic analyses were conducted to clarify evolutionary relationships with other PKoV strains.
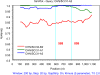
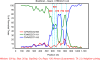
Results: Approximately 53% (87/163) of pigs tested positive for PKoV. PKoV was widespread in asymptomatic pigs and those with diarrhea. A high prevalence of PKoV was observed in pigs younger than 4 weeks and in pigs with diarrhea. Phylogenetic analysis of 36 PKoV VP1 protein sequences showed that Sichuan PKoV strains formed four distinct clusters. Two pigs with diarrhea were found to be co-infected with multiple PKoV strains. Sequence and phylogenetic analyses revealed diversity within the same host and between different hosts. Significant recombination breakpoints were observed between the CHN/SC/31-A1 and CHN/SC/31-A3 strains in the VP1 region, which were isolated from the same sample.
Conclusion: PKoV was endemic in Sichuan Province regardless of whether pigs were healthy or suffering from diarrhea. Based on our statistical analyses, we suggest that PKoV was the likely causative agent of high-mortality diarrhea in China from 2010. For the first time, we provide evidence for the co-existence of multiple PKoV strains in one pig, and possible recombination events in the VP1 region. Our findings provide further insights into the molecular properties of PKoV, along with its epidemiology.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

