Migrating bubble during break-induced replication drives conservative DNA synthesis
- PMID: 24025772
- PMCID: PMC3804423
- DOI: 10.1038/nature12584
Migrating bubble during break-induced replication drives conservative DNA synthesis
Abstract
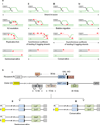
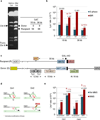
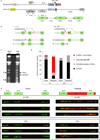
The repair of chromosomal double strand breaks (DSBs) is crucial for the maintenance of genomic integrity. However, the repair of DSBs can also destabilize the genome by causing mutations and chromosomal rearrangements, the driving forces for carcinogenesis and hereditary diseases. Break-induced replication (BIR) is one of the DSB repair pathways that is highly prone to genetic instability. BIR proceeds by invasion of one broken end into a homologous DNA sequence followed by replication that can copy hundreds of kilobases of DNA from a donor molecule all the way through its telomere. The resulting repaired chromosome comes at a great cost to the cell, as BIR promotes mutagenesis, loss of heterozygosity, translocations, and copy number variations, all hallmarks of carcinogenesis. BIR uses most known replication proteins to copy large portions of DNA, similar to S-phase replication. It has therefore been suggested that BIR proceeds by semiconservative replication; however, the model of a bona fide, stable replication fork contradicts the known instabilities associated with BIR such as a 1,000-fold increase in mutation rate compared to normal replication. Here we demonstrate that in budding yeast the mechanism of replication during BIR is significantly different from S-phase replication, as it proceeds via an unusual bubble-like replication fork that results in conservative inheritance of the new genetic material. We provide evidence that this atypical mode of DNA replication, dependent on Pif1 helicase, is responsible for the marked increase in BIR-associated mutations. We propose that the BIR mode of synthesis presents a powerful mechanism that can initiate bursts of genetic instability in eukaryotes, including humans.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

