Nkx genes are essential for maintenance of ventricular identity
- PMID: 24026123
- PMCID: PMC3787760
- DOI: 10.1242/dev.095562
Nkx genes are essential for maintenance of ventricular identity
Abstract
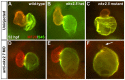
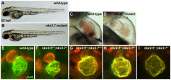
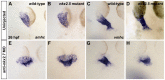
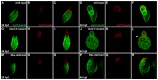
Establishment of specific characteristics of each embryonic cardiac chamber is crucial for development of a fully functional adult heart. Despite the importance of defining and maintaining unique features in ventricular and atrial cardiomyocytes, the regulatory mechanisms guiding these processes are poorly understood. Here, we show that the homeodomain transcription factors Nkx2.5 and Nkx2.7 are necessary to sustain ventricular chamber attributes through repression of atrial chamber identity. Mutation of nkx2.5 in zebrafish yields embryos with diminutive ventricular and bulbous atrial chambers. These chamber deformities emerge gradually during development, with a severe collapse in the number of ventricular cardiomyocytes and an accumulation of excess atrial cardiomyocytes as the heart matures. Removal of nkx2.7 function from nkx2.5 mutants exacerbates the loss of ventricular cells and the gain of atrial cells. Moreover, in these Nkx-deficient embryos, expression of vmhc, a ventricular gene, fades, whereas expression of amhc, an atrial gene, expands. Cell-labeling experiments suggest that ventricular cardiomyocytes can transform into atrial cardiomyocytes in the absence of Nkx gene function. Through suggestion of transdifferentiation from ventricular to atrial fate, our data reveal a pivotal role for Nkx genes in maintaining ventricular identity and highlight remarkable plasticity in differentiated myocardium. Thus, our results are relevant to the etiologies of fetal and neonatal cardiac pathology and could direct future innovations in cardiac regenerative medicine.
Keywords: Atrium; Chamber identity; Ventricle; Zebrafish; nkx2.5; nkx2.7.
Figures





References
-
- Alexander J., Stainier D. Y., Yelon D. (1998). Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev. Genet. 22, 288–299 - PubMed
-
- Azpiazu N., Frasch M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7 7B, 1325–1340 - PubMed
-
- Bao Z. Z., Bruneau B. G., Seidman J. G., Seidman C. E., Cepko C. L. (1999). Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283, 1161–1164 - PubMed
-
- Benson D. W., Silberbach G. M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S., Smalls O., Johnson M. C., Watson M. S., Seidman J. G., et al. (1999). Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Invest. 104, 1567–1573 - PMC - PubMed
-
- Berdougo E., Coleman H., Lee D. H., Stainier D. Y., Yelon D. (2003). Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130, 6121–6129 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

