Analysis of variable retroduplications in human populations suggests coupling of retrotransposition to cell division
- PMID: 24026178
- PMCID: PMC3847774
- DOI: 10.1101/gr.154625.113
Analysis of variable retroduplications in human populations suggests coupling of retrotransposition to cell division
Abstract
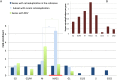
In primates and other animals, reverse transcription of mRNA followed by genomic integration creates retroduplications. Expressed retroduplications are either "retrogenes" coding for functioning proteins, or expressed "processed pseudogenes," which can function as noncoding RNAs. To date, little is known about the variation in retroduplications in terms of their presence or absence across individuals in the human population. We have developed new methodologies that allow us to identify "novel" retroduplications (i.e., those not present in the reference genome), to find their insertion points, and to genotype them. Using these methods, we catalogued and analyzed 174 retroduplication variants in almost one thousand humans, which were sequenced as part of Phase 1 of The 1000 Genomes Project Consortium. The accuracy of our data set was corroborated by (1) multiple lines of sequencing evidence for retroduplication (e.g., depth of coverage in exons vs. introns), (2) experimental validation, and (3) the fact that we can reconstruct a correct phylogenetic tree of human subpopulations based solely on retroduplications. We also show that parent genes of retroduplication variants tend to be expressed at the M-to-G1 transition in the cell cycle and that M-to-G1 expressed genes have more copies of fixed retroduplications than genes expressed at other times. These findings suggest that cell division is coupled to retrotransposition and, perhaps, is even a requirement for it.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
