Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses
- PMID: 24027312
- PMCID: PMC3807891
- DOI: 10.1128/JVI.02013-13
Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses
Abstract
Bats are considered important animal reservoirs for many viruses pathogenic to humans. An approach based on viral metagenomics was used to study gut specimens from 78 insectivorous bats in Yunnan Province, China. Seventy-four reads were found to be related to group A rotavirus (RVA). Further reverse transcription-PCR screening and viral isolation on cell cultures confirmed the presence of a novel RVA strain, named RVA/Bat-tc/MSLH14/2012/G3P[3], in 1 (6%) of 16 lesser horseshoe bats. Full genomic sequencing analyses showed that MSLH14 possessed the genotype constellation G3-P[3]-I8-R3-C3-M3-A9-N3-T3-E3-H6, which is akin to human and animal rotaviruses believed to be of feline/canine origin. Phylogenetic analysis indicated that VP7 was most closely related to bovine RVA strains from India, whereas VP4 was most closely related to an unusual human RVA strain, CMH222, with animal characteristics isolated in Thailand. The remaining gene segments were only distantly related to a range of animal RVA strains, most of which are believed to be related to feline/canine RVAs. Experimental infection showed that bat RVA strain MSLH14 was highly pathogenic to suckling mice, causing 100% mortality when they were inoculated orally with a titer as low as 5 × 10² 50% tissue culture infective doses. As this virus is not closely related to any known RVA strain, it is tempting to speculate that it is a true bat RVA strain rather than a virus transmitted between species. However, further screening of bat populations, preferably juvenile animals, will be crucial in determining whether or not this virus is widely distributed in the bat population.
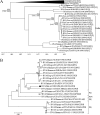
Figures





References
-
- Estes MK, Kapikian AZ. 2007. Rotaviruses, p. 1917–1974 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA
-
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, WHO-Coordinated Global Rotavirus Surveillance Network 2012. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 12:136–141 - PubMed
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Rotavirus, p 603–613 In Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA
-
- Matthijnssens J, Van Ranst M. 2012. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2:426–433 - PubMed
-
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204–3219 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

