Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles
- PMID: 24027323
- PMCID: PMC3838225
- DOI: 10.1128/JVI.00704-13
Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles
Abstract
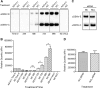
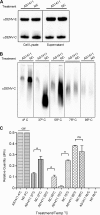
Dengue virus (DENV) is a mosquito-transmitted flavivirus that can cause severe disease in humans and is considered a reemerging pathogen of significant importance to public health. The DENV capsid (C) protein functions as a structural component of the infectious virion; however, it may have additional functions in the virus replicative cycle. Here, we show that the DENV C protein interacts and colocalizes with the multifunctional host protein nucleolin (NCL). Furthermore, we demonstrate that this interaction can be disrupted by the addition of an NCL binding aptamer (AS1411). Knockdown of NCL with small interfering RNA (siRNA) or treatment of cells with AS1411 results in a significant reduction of viral titers after DENV infection. Western blotting and quantitative RT-PCR (qRT-PCR) analysis revealed no differences in viral RNA or protein levels at early time points postinfection, suggesting a role for NCL in viral morphogenesis. We support this hypothesis by showing that treatment with AS1411 alters the migration characteristics of the viral capsid, as visualized by native electrophoresis. Here, we identify a critical interaction between DENV C protein and NCL that represents a potential new target for the development of antiviral therapeutics.
Figures

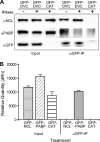
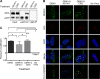

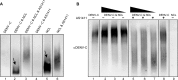

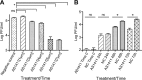

References
-
- WHO 2006. Scientific working group report on dengue. WHO reference number TDR/SWG/08. WHO, Geneva, Switzerland
-
- Kyle JL, Harris E. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 62:71–92 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

