A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA
- PMID: 24027348
- PMCID: PMC3734968
- DOI: 10.3767/003158513X666268
A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA
Abstract
The basal fungal order Mortierellales constitutes one of the largest orders in the basal lineages. This group consists of one family and six genera. Most species are saprobic soil inhabiting fungi with the ability of diverse biotransformations or the accumulation of unsaturated fatty acids, making them attractive for biotechnological applications. Only few studies exist aiming at the revelation of the evolutionary relationships of this interesting fungal group. This study includes the largest dataset of LSU and ITS sequences for more than 400 specimens containing 63 type or reference strains. Based on a LSU phylogram, fungal groups were defined and evaluated using ITS sequences and morphological features. Traditional morphology-based classification schemes were rejected, because the morphology of the Mortierellales seems to depend on culture conditions, a fact, which makes the identification of synapomorphic characters tedious. This study belongs to the most comprehensive molecular phylogenetic analyses for the Mortierellales up to date and reveals unresolved species and species complexes.
Keywords: Zygomycetes; Zygomycota; internal transcribed spacer; large subunit ribosomal DNA; taxonomic revision.
Figures
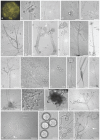




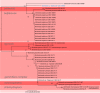
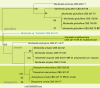


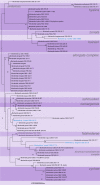
References
-
- Cavalier-Smith T.1998. A revised six-kingdom system of life. Biological Reviews of the Cambridge Philosophical Society 73, 3: 203–266 - PubMed
-
- Chalabuda TV.1968. Systemica familiae Mortierella. Novosti Sistematiki Nizshikh Rastenii 5: 120–131
-
- Chesters CGC.1933. Azygozygum chlamydosporum nov. gen. et sp. A phycomycete associated with a diseased condition of Antirrhinum majus. Transactions of the British Mycological Society 18, 3: 199–214
-
- Coemans E.1863. Quelques hyphomycetes nouveaux. 1. Mortierella polycephala et Martensella pectinata. Bulletin de l’Académie Royale des Sciences de Belgique Classe des Sciences 2, ser. 15: 536–544
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
