MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer
- PMID: 24027433
- PMCID: PMC3769887
- DOI: 10.1593/neo.13998
MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer
Abstract
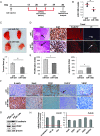
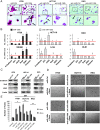
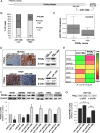
MicroRNA-130b (miR-130b) is involved in several biologic processes; its role in colorectal tumorigenesis has not been addressed so far. Herein, we demonstrate that miR-130b up-regulation exhibits clinical relevance as it is linked to advanced colorectal cancers (CRCs), poor patients' prognosis, and molecular features of enhanced epithelial-mesenchymal transition (EMT) and angiogenesis. miR-130b high-expressing cells develop large, dedifferentiated, and vascularized tumors in mouse xenografts, features that are reverted by intratumor injection of a specific antisense RNA. In contrast, injection of the corresponding mimic in mouse xenografts from miR-130b low-expressing cells increases tumor growth and angiogenic potential while reduces the epithelial hallmarks. These biologic effects are reproduced in human CRC cell lines. We identify peroxisome proliferator-activated receptor γ (PPARγ) as an miR-130b direct target in CRC in vitro and in vivo. Notably, the effects of PPARγ gain- and loss-of-function phenocopy those due to miR-130b down-regulation or up-regulation, respectively, underscoring their biologic relevance. Furthermore, we provide mechanistic evidences that most of the miR-130b-dependent effects are due to PPARγ suppression that in turn deregulates PTEN, E-cadherin, Snail, and vascular endothelial growth factor, key mediators of cell proliferation, EMT, and angiogenesis. Since higher levels of miR-130b are found in advanced tumor stages (III-IV), we propose a novel role of the miR-130b-PPARγ axis in fostering the progression toward more invasive CRCs. Detection of onco-miR-130b and its association with PPARγ may be useful as a prognostic biomarker. Its targeting in vivo should be evaluated as a novel effective therapeutic tool against CRC.
Figures



References
-
- Dean M. Cancer as a complex developmental disorder—nineteenth Cornelius P. Rhoads Memorial Award Lecture. Cancer Res. 1998;58:5633–5636. - PubMed
-
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. - PubMed
-
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
