Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice
- PMID: 24030380
- PMCID: PMC3814730
- DOI: 10.1182/blood-2013-06-508432
Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice
Abstract
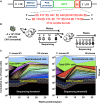
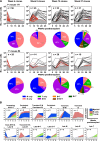
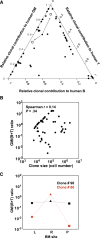
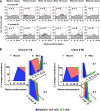
Human cord blood (CB) offers an attractive source of cells for clinical transplants because of its rich content of cells with sustained repopulating ability in spite of an apparent deficiency of cells with rapid reconstituting ability. Nevertheless, the clonal dynamics of nonlimiting CB transplants remain poorly understood. To begin to address this question, we exposed CD34+ CB cells to a library of barcoded lentiviruses and used massively parallel sequencing to quantify the clonal distributions of lymphoid and myeloid cells subsequently detected in sequential marrow aspirates obtained from 2 primary NOD/SCID-IL2Rγ(-/-) mice, each transplanted with ∼10(5) of these cells, and for another 6 months in 2 secondary recipients. Of the 196 clones identified, 68 were detected at 4 weeks posttransplant and were often lympho-myeloid. The rest were detected later, after variable periods up to 13 months posttransplant, but with generally increasing stability throughout time, and they included clones in which different lineages were detected. However, definitive evidence of individual cells capable of generating T-, B-, and myeloid cells, for over a year, and self-renewal of this potential was also obtained. These findings highlight the caveats and utility of this model to analyze human hematopoietic stem cell control in vivo.
Figures
References
-
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–136. - PubMed
-
- Muller-Sieburg CE, Sieburg HB. Clonal diversity of the stem cell compartment. Curr Opin Hematol. 2006;13(4):243–248. - PubMed
-
- Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4(2):220–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

