Identification of novel markers for mouse CD4(+) T follicular helper cells
- PMID: 24030473
- PMCID: PMC3947211
- DOI: 10.1002/eji.201343469
Identification of novel markers for mouse CD4(+) T follicular helper cells
Erratum in
- Eur J Immunol. 2014 Jul;44(7):2197
Abstract
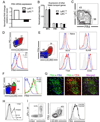
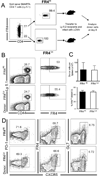
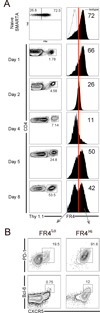
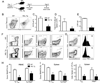
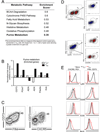
CD4(+) T follicular helper (TFH) cells are central for generation of long-term B-cell immunity. A defining phenotypic attribute of TFH cells is the expression of the chemokine R CXCR5, and TFH cells are typically identified by co-expression of CXCR5 together with other markers such as PD-1, ICOS, and Bcl-6. Herein, we report high-level expression of the nutrient transporter folate R 4 (FR4) on TFH cells in acute viral infection. Distinct from the expression profile of conventional TFH markers, FR4 was highly expressed by naive CD4(+) T cells, was downregulated after activation and subsequently re-expressed on TFH cells. Furthermore, FR4 expression was maintained, albeit at lower levels, on memory TFH cells. Comparative gene expression profiling of FR4(hi) versus FR4(lo) Ag-specific CD4(+) effector T cells revealed a molecular signature consistent with TFH and TH1 subsets, respectively. Interestingly, genes involved in the purine metabolic pathway, including the ecto-enzyme CD73, were enriched in TFH cells compared with TH1 cells, and phenotypic analysis confirmed expression of CD73 on TFH cells. As there is now considerable interest in developing vaccines that would induce optimal TFH cell responses, the identification of two novel cell surface markers should be useful in characterization and identification of TFH cells following vaccination and infection.
Keywords: B cells; CD4+ T cells; CD73; Folate R 4; Viral infection.
© 2013 The Authors. European Journal of Immunology published by Wiley‐VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have no conflicts of interest to disclose
Figures


References
-
- Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121–1129. - PubMed
-
- Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

