Characterization of class 1 integrons and antibiotic resistance genes in multidrug-resistant Salmonella enterica isolates from foodstuff and related sources
- PMID: 24031680
- PMCID: PMC3769850
- DOI: 10.1590/S1517-838220110002000033
Characterization of class 1 integrons and antibiotic resistance genes in multidrug-resistant Salmonella enterica isolates from foodstuff and related sources
Abstract
In recent years, an increase in the occurrence of antimicrobial resistance among Salmonella enterica has been observed in several countries, which is worrisome because S. enterica is one of the most common causes of human gastroenteritis worldwide. The aim of this study was to characterize class 1 integrons and antibiotic resistance genotypes in Salmonella enterica isolates recovered from foodstuff and related sources. Nineteen multidrug-resistant (MDR) Salmonella enterica isolates were recovered. Higher resistance rates to tetracycline (90%), streptomycin (80%), sulfamethoxazole-trimethoprim (80%), ampicillin (60%) and nalidixic acid (70%) were related to the presence of the tetA, aadA, sul1/sul2, bla TEM-1 genes, and a codon mutation at position 83 of the gyrA gene, respectively. Class 1 integrons harboring aadA, bla TEM-1, sul1 or dhfr1 genes were detected in nine (45%) Salmonella enterica strains belonging to serotypes Brandenburg, Panama, Agona, Mbandaka and Alachua. Finally, clonal dissemination of S. Panama, S. Derby and S. Mbandaka was confirmed by PFGE. Detection of clonally related MDR Salmonella enterica suggests that endemic serotypes can be supported by class 1 integron-borne gene cassettes and/or mutations in drug targets. Emergence and dissemination of multidrug-resistant Salmonella enterica can have a major public health impact in an environment where large-scale suppliers ship their products.
Keywords: Brazil; Salmonella; class 1 integrons; foodstuff; multidrug-resistant.
Figures
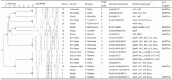
No repeated samples were included; salami samples were taken after curing.
Susceptible to all antibiotics tested.
Amp (ampicillin), Cm (chloramphenicol), Enr (enrofloxacin), Gm (gentamicin), Km (kanamycin), Nal (nalidixic acid), Sxt (sulfamethoxazole/trimethoprim), Sm (streptomycin), Tc (tetracycline).
Ser-83-Phe: Substitution of Serine for Phenylalanine in the codon 83 of the gyrA gene.
[class 1 integron]; [aadA, qacE_I, sul1], 1700 bp; [dhfr], 700 bp; [aadA, dhfr], 1700 bp; [aadA, blaTEM, qacE_I ,sul1, sul2], 2700 bp.
References
-
- Amar C.F., Arnold C., Bankier A., Dear P.H., Guerra B., Hopkins K.L., Liebana E., Mevius D.J., Threlfall E.J. Real-time PCRs and fingerprinting assays for the detection and characterization of Salmonella Genomic Island-1 encoding multidrug resistance: application to 445 European isolates of Salmonella, Escherichia coli, Shigella, and Proteus. Microb. Drug. Resist. 2008;14(2):79–92. - PubMed
-
- Antunes P., Machado J., Peixe L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J. Antimicrob. Chemother. 2006;58(2):297–304. - PubMed
-
- Arcangioli M.A., Leroy-Setrin S., Martel J.L., Chaslus-Dancla E. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J. Med. Microbiol. 2000;49(1):103–110. - PubMed
-
- Carramiñana J.J., Rota C., Agustín I., Herrera A. High prevalence of multiple resistance to antibiotics in Salmonella serovars isolated from a poultry slaughterhouse in Spain. Vet. Microbiol. 2004;104(1–2):133–139. - PubMed
-
- Centers for Disease Control {homepage on the internet}. Atlanta: One-day (24–28h) standardized laboratory protocol for molecular subtyping of Salmonella serotype Typhimurium by pulsed field gel electrophoresis (PFGE) 1999. {updated 2009 Apr 29; cited 2010 Jan 10} PulseNet: The national molecular subtyping network for foodborne disease surveillance, Section 5.2; {about 2 screens}. Available from: http://www.cdc.gov/pulsenet.
LinkOut - more resources
Full Text Sources
Miscellaneous
