Reconciliation revisited: handling multiple optima when reconciling with duplication, transfer, and loss
- PMID: 24033262
- PMCID: PMC3791060
- DOI: 10.1089/cmb.2013.0073
Reconciliation revisited: handling multiple optima when reconciling with duplication, transfer, and loss
Abstract
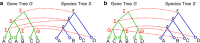
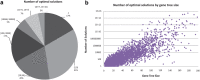
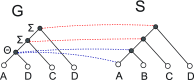
Phylogenetic tree reconciliation is a powerful approach for inferring evolutionary events like gene duplication, horizontal gene transfer, and gene loss, which are fundamental to our understanding of molecular evolution. While duplication-loss (DL) reconciliation leads to a unique maximum-parsimony solution, duplication-transfer-loss (DTL) reconciliation yields a multitude of optimal solutions, making it difficult to infer the true evolutionary history of the gene family. This problem is further exacerbated by the fact that different event cost assignments yield different sets of optimal reconciliations. Here, we present an effective, efficient, and scalable method for dealing with these fundamental problems in DTL reconciliation. Our approach works by sampling the space of optimal reconciliations uniformly at random and aggregating the results. We show that even gene trees with only a few dozen genes often have millions of optimal reconciliations and present an algorithm to efficiently sample the space of optimal reconciliations uniformly at random in O(mn(2)) time per sample, where m and n denote the number of genes and species, respectively. We use these samples to understand how different optimal reconciliations vary in their node mappings and event assignments and to investigate the impact of varying event costs. We apply our method to a biological dataset of approximately 4700 gene trees from 100 taxa and observe that 93% of event assignments and 73% of mappings remain consistent across different multiple optima. Our analysis represents the first systematic investigation of the space of optimal DTL reconciliations and has many important implications for the study of gene family evolution.
Figures


References
-
- Bansal M.S. Burleigh J.G. Eulenstein O. Wehe A. Heuristics for the gene-duplication problem: A Θ(n) speed-up for the local search. RECOMB. 2007:238–252.
-
- Bonizzoni P. Vedova G.D. Dondi R. Reconciling a gene tree to a species tree under the duplication cost model. Theor. Comput. Sci. 2005;347:36–53.
-
- Charleston M. Jungles: A new solution to the host-parasite phylogeny reconciliation problem. Mathematical Biosciences. 1998;149:191–223. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

