Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization
- PMID: 24035354
- PMCID: PMC3825755
- DOI: 10.1016/j.stem.2013.08.013
Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization
Abstract
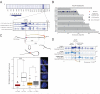
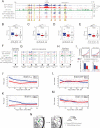
The relationship between 3D organization of the genome and gene-regulatory networks is poorly understood. Here, we examined long-range chromatin interactions genome-wide in mouse embryonic stem cells (ESCs), iPSCs, and fibroblasts and uncovered a pluripotency-specific genome organization that is gradually reestablished during reprogramming. Our data confirm that long-range chromatin interactions are primarily associated with the spatial segregation of open and closed chromatin, defining overall chromosome conformation. Additionally, we identified two further levels of genome organization in ESCs characterized by colocalization of regions with high pluripotency factor occupancy and strong enrichment for Polycomb proteins/H3K27me3, respectively. Underlining the independence of these networks and their functional relevance for genome organization, loss of the Polycomb protein Eed diminishes interactions between Polycomb-regulated regions without altering overarching chromosome conformation. Together, our data highlight a pluripotency-specific genome organization in which pluripotency factors such as Nanog and H3K27me3 occupy distinct nuclear spaces and reveal a role for cell-type-specific gene-regulatory networks in genome organization.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
