The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance
- PMID: 24036343
- PMCID: PMC3837940
- DOI: 10.1128/EC.00103-13
The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance
Abstract
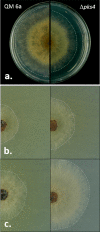
Species of the fungal genus Trichoderma (Hypocreales, Ascomycota) are well-known for their production of various secondary metabolites. Nonribosomal peptides and polyketides represent a major portion of these products. In a recent phylogenomic investigation of Trichoderma polyketide synthase (PKS)-encoding genes, the pks4 from T. reesei was shown to be an orthologue of pigment-forming PKSs involved in synthesis of aurofusarin and bikaverin in Fusarium spp. In this study, we show that deletion of this gene in T. reesei results in loss of green conidial pigmentation and in pigmentation alteration of teleomorph structures. It also has an impact on conidial cell wall stability and the antagonistic abilities of T. reesei against other fungi, including formation of inhibitory metabolites. In addition, deletion of pks4 significantly influences the expression of other PKS-encoding genes of T. reesei. To our knowledge, this is the first indication that a low-molecular-weight pigment-forming PKS is involved in defense, mechanical stability, and stress resistance in fungi.
Figures




References
-
- Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. 2011. Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 16:749–759 - PubMed
-
- Druzhinina IS, Kubicek CP. 2013. Ecological genomics of Trichoderma. In Martin F. (ed), Ecological genomics of fungi. Wiley-Blackwell, Oxford, United Kingdom
-
- Mukherjee PK, Horwitz BA, Kenerley CM. 2012. Secondary metabolism in Trichoderma: a genomic perspective. Microbiology 158:35–45 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

