Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration
- PMID: 24036952
- PMCID: PMC3902040
- DOI: 10.1038/ng.2741
Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration
Abstract
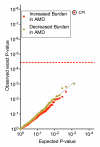
To define the role of rare variants in advanced age-related macular degeneration (AMD) risk, we sequenced the exons of 681 genes within all reported AMD loci and related pathways in 2,493 cases and controls. We first tested each gene for increased or decreased burden of rare variants in cases compared to controls. We found that 7.8% of AMD cases compared to 2.3% of controls are carriers of rare missense CFI variants (odds ratio (OR) = 3.6; P = 2 × 10(-8)). There was a predominance of dysfunctional variants in cases compared to controls. We then tested individual variants for association with disease. We observed significant association with rare missense alleles in genes other than CFI. Genotyping in 5,115 independent samples confirmed associations with AMD of an allele in C3 encoding p.Lys155Gln (replication P = 3.5 × 10(-5), OR = 2.8; joint P = 5.2 × 10(-9), OR = 3.8) and an allele in C9 encoding p.Pro167Ser (replication P = 2.4 × 10(-5), OR = 2.2; joint P = 6.5 × 10(-7), OR = 2.2). Finally, we show that the allele of C3 encoding Gln155 results in resistance to proteolytic inactivation by CFH and CFI. These results implicate loss of C3 protein regulation and excessive alternative complement activation in AMD pathogenesis, thus informing both the direction of effect and mechanistic underpinnings of this disorder.
Figures



References
-
- Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379:1728–1738. - PubMed
-
- Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. - PubMed
-
- Friedman DS, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

