Protein evolution along phylogenetic histories under structurally constrained substitution models
- PMID: 24037213
- PMCID: PMC5221705
- DOI: 10.1093/bioinformatics/btt530
Protein evolution along phylogenetic histories under structurally constrained substitution models
Abstract
Motivation: Models of molecular evolution aim at describing the evolutionary processes at the molecular level. However, current models rarely incorporate information from protein structure. Conversely, structure-based models of protein evolution have not been commonly applied to simulate sequence evolution in a phylogenetic framework, and they often ignore relevant evolutionary processes such as recombination. A simulation evolutionary framework that integrates substitution models that account for protein structure stability should be able to generate more realistic in silico evolved proteins for a variety of purposes.
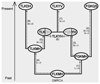
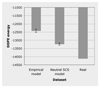
Results: We developed a method to simulate protein evolution that combines models of protein folding stability, such that the fitness depends on the stability of the native state both with respect to unfolding and misfolding, with phylogenetic histories that can be either specified by the user or simulated with the coalescent under complex evolutionary scenarios, including recombination, demographics and migration. We have implemented this framework in a computer program called ProteinEvolver. Remarkably, comparing these models with empirical amino acid replacement models, we found that the former produce amino acid distributions closer to distributions observed in real protein families, and proteins that are predicted to be more stable. Therefore, we conclude that evolutionary models that consider protein stability and realistic evolutionary histories constitute a better approximation of the real evolutionary process.
Figures
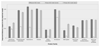
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

