MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells
- PMID: 24037508
- PMCID: PMC3955211
- DOI: 10.1038/nsmb.2665
MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells
Abstract
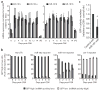
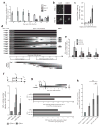
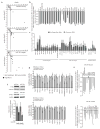
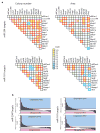
Individual microRNAs (miRNAs) can target hundreds of mRNAs forming networks of presumably cooperating genes. To test this presumption, we functionally screened miRNAs and their targets in the context of dedifferentiation of mouse fibroblasts to induced pluripotent stem cells (iPSCs). Along with the miR-302-miR-294 family, the miR-181 family arose as a previously unidentified enhancer of the initiation phase of reprogramming. Endogenous miR-181 miRNAs were transiently elevated with the introduction of Pou5f1 (also known as Oct4), Sox2 and Klf4 (referred to as OSK), and miR-181 inhibition diminished iPSC colony formation. We tested the functional contribution of 114 individual targets of the two families, revealing 25 genes that normally suppress initiation. Coinhibition of targets cooperatively promoted both the frequency and kinetics of OSK-induced reprogramming. These data establish two of the largest functionally defined networks of miRNA-mRNA interactions and reveal previously unidentified relationships among genes that act together to suppress early stages of reprogramming.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

