Genome-wide identification and functional analyses of microRNA signatures associated with cancer pain
- PMID: 24039159
- PMCID: PMC3840489
- DOI: 10.1002/emmm.201302797
Genome-wide identification and functional analyses of microRNA signatures associated with cancer pain
Abstract
Cancer pain remains a major challenge and there is an urgent demand for the development of specific mechanism-based therapies. Various diseases are associated with unique signatures of expression of microRNAs (miRNAs), which reveal deep insights into disease pathology. Using a comprehensive approach combining genome-wide miRNA screening, molecular and in silico analyses with behavioural approaches in a clinically relevant model of metastatic bone-cancer pain in mice, we now show that tumour-induced conditions are associated with a marked dysregulation of 57 miRNAs in sensory neurons corresponding to tumour-affected areas. By establishing protocols for interference with disease-induced miRNA dysregulation in peripheral sensory neurons in vivo, we functionally validate six dysregulated miRNAs as significant modulators of tumour-associated hypersensitivity. In silico analyses revealed that their predicted targets include key pain-related genes and we identified Clcn3, a gene encoding a chloride channel, as a key miRNA target in sensory neurons, which is functionally important in tumour-induced nociceptive hypersensitivity in vivo. Our results provide new insights into endogenous gene regulatory mechanisms in cancer pain and open up attractive and viable therapeutic options.
Keywords: Clcn3; bone metastatic pain; gene regulation; miRNA inhibitors; miRNA mimics.
© 2013 The Authors. Published by John Wiley and Sons, Ltd on behalf of EMBO.
Figures

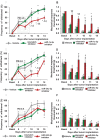
A. Increase in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice as compared to sham surgery. * denotes p = 0.002 on PID-5, 6, 7 and <0.0001 from PID-8 through 15 as compared to basal and † denotes p < 0.001 on PID-5 and <0.0001 from PID-6 through 15 as compared to corresponding data point in the sham group, two-way ANOVA of repeated measures followed by Bonferroni's multiple comparisons post hoc test, n = at least 6 mice per group.
B. Mechanical response threshold calculated as von Frey filament strength required to achieve 50% withdrawal frequency. * denotes p < 0.001 from PID-4 through 15 as compared to basal and † denotes p = 0.004 on PID-5, 6 & 13, 0.006 on PID-7, 9 & 11, 0.005 on PID-8, 0.004 on PID-10, 0.0001 on PID-12 & 14, and 0.003 on PID-15 as compared to corresponding data point in the sham group, two-way ANOVA of repeated measures followed by Bonferroni's multiple comparisons post hoc test, n = at least 6 mice per group.
C,D. Heat maps of miRNAs found to be significantly up- or downregulated via microarray analysis in the ipsilateral lumbar DRG of tumor-bearing mice 4 days (C) or 8 days (D) post implantation as compared to sham surgery. Scale indicates expression intensities obtained from the microarray experiment.
E. Representation of examples of miRNAs showing up- or down-regulation following independent verification with quantitative RT-PCR analyses (left hand panel) and the original data from microarray analysis. *p = 0.001 for miR-544-3p, 0.003 for miR-1a-3p, 0.009 for miR-34c-5p, 0.04 for miR-370-3p, 0.03 for miR-291b-5p and 0.005 for miR-483-3p as compared to sham-treated group, ANOVA followed by post hoc Fischer's test, n = 3 mice per group.

Experimental scheme established in this study which enables effective knockdown/induction of miRNA expression in DRGs in vivo and analysis of tumour pain-associated behaviours.
Microscopic analyses of whole-mount DRGs or cryosections showing uptake of FAM-conjugated miRNA inhibitors. Scale bar is 50 µm in all panels.
Typical examples of qRT-PCR verification of efficacy of miRNA inhibitors in reversing tumour-induced upregulation of miRNAs in ipsilateral DRGs in vivo.
Typical examples of qRT-PCR verification of efficacy of miRNA mimics in reversing tumour-induced downregulation of miRNAs and inducing overexpression of miRNAs in ipsilateral DRGs in vivo. In panel (C), * denotes p = 0.02 for miR-1a-3p, 0.04 for miR-34c-3p as compared to corresponding mismatch inhibitor and in panel (D), * denotes p = 0.001 for miR-370-3p and <0.0001 for miR-291b-5p as compared to non-targeting mimic, ANOVA followed by post hoc Fischer's test, n = 3 per group.
Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-1a-3p inhibitor (red symbols) or the corresponding mismatch inhibitors (green symbols) or vehicle (grey symbols). * denotes p < 0.0001 on PID-5 through 15 in the vehicle, mismatch inhibitor and miR-1a-3p inhibitor groups as compared to basal; † denotes p = 0.007 on PID-10 and <0.0001 on PID-12 & 14 as compared to corresponding data point in the mismatch inhibitor group; ‡ denotes p < 0.0001 on PID-10, 12 & 14 as compared to corresponding data point in the vehicle group.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 50% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-1a-3p inhibitor (red bars) or the corresponding mismatch inhibitors (green bars) or vehicle (grey bars). * denotes p < 0.0001 from PID-5 through 14 in the vehicle and mismatch inhibitor groups and on PID-10, 12, 14 in the miR-1a-3p-inhibitor groupas compared to basal; † denotes p = 0.004 on PID-5, 0.003 on PID-7, 0.05 on PID-10 and 0.01 on PID-12 as compared to corresponding data point in the mismatch inhibitor group; ‡ denotes p < 0.0001 on PID-5, 0.0002 on PID-7, 0.05 on PID-10, and 0.004 on PID-12 as compared to corresponding data point in the vehicle group.
Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-34c-5p inhibitor (red symbols) or the corresponding mismatch inhibitor (green symbols) or vehicle (grey symbols). * denotes p < 0.0001 on PID-5 through 14 in the vehicle and mismatch inhibitor groups, 0.006 on PID-5 and < 0.0001 on PID-14 for miR-34c-5p- inhibitor group as compared to basal; † denotes p < 0.0001 on PID-7, 9 & 11 as compared to corresponding data point in the mismatch inhibitor group; ‡ denotes p < 0.0001 on PID-7, 9 & 11 as compared to corresponding data point in the vehicle group.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 50% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-34c-5p inhibitor (red bars) or the corresponding mismatch inhibitors (green bars) or vehicle (grey bars). * denotes p = 0.0003 on PID-7 & 9, < 0.0001 on PID-11 & 14 in the mismatch-inhibitor group; 0.0018 on PID-5 and < 0.0001 on PID-14 in the miR-34c-5p-inhibitor group and 0.0001 on PID-5 through 15 in the vehicle group; † denotes p < 0.0001 on PID-7, 0.0009 on PID-9, 0.05 on PID-11 as compared to corresponding data point in the mismatch inhibitor group; and ‡ denotes p < 0.0001 on PID-7 & 9, 0.0518 on PID-11 as compared to corresponding data point in the vehicle group.
Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-544-3p inhibitor (red symbols) or the corresponding mismatch inhibitors (green symbols) or vehicle (grey symbols). * denotes p < 0.0001 from PID-3 through PID-14 in vehicle, mismatch-inhibitor and miR-544-3p inhibitor groups.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 50% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-544-3p inhibitor (red bars) or the corresponding mismatch inhibitors (green bars) or vehicle (grey bars). * denotes p < 0.0001 from PID-3 through PID-14 in mismatch-inhibitor and miR-544-3p inhibitor groups.

Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.02 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-370-3p mimic (red symbols) or non-targeting mimic (green) or vehicle (grey symbols). * denotes p = 0.0192 on PID-13 in the vehicle group and <0.0001 on PID-8 through 15 in the miR-370-3p-mimic group as compared to basal; † denotes p = 0.0013 on PID-8 and <0.001 on PID-10, 13 & 15 as compared to corresponding data point in the non-targeting mimic group; ‡ denotes p = 0.0003 on PID-8, <0.0001 on PID-10 & 15 and 0.0044 on PID-13 as compared to corresponding data point in the vehicle group.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 80% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally deliveredmiR-370-3p mimic (red bars) or non-targeting mimic (green bars) or vehicle (grey bars). * denotes p = 0.0291 on PID-8, 0.05 on PID-10, 0.0192 on PID-13 and 0.05 on PID-15 in the vehicle group, 0.04 on PID-8, 0.002 on PID-10, 0.05 on PID-13 & 15 in the non-targeting mimic group; † denotes p = 0.0013 on PID-3, <0.0001 on PID-6, 8 & 10 as compared to corresponding data point in the non-targeting mimic group; ‡ denotes p = 0.0013 on PID-6 and <0.0001 on PID-8, 10 & 13 as compared to corresponding data point in the vehicle group.
Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-483-3p mimic (red symbols) or non-targeting mimic (green) or vehicle (grey symbols). * denotes p = 0.0008 on PID-6, 0.0002 on PID-8 & <0.0001 on PID-10, 13, 15 in vehicle, non-targeting mimic and miR-483-3p mimic groups as compared to basal; † denotes p = 0.0206 on PID-10 & 0.0074 on PID-13 as compared to corresponding data point in the non-targeting mimic group; and ‡ denotes p = 0.0074 on PID-13 as compared to corresponding data point in the vehicle group.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 50% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally deliveredmiR-483-3p mimic (red bars) or non-targeting mimic (green bars) or vehicle (grey bars). * denotes p < 0.0001 from PID-3 through 15 in vehicle, non-targeting-mimic groups and miR-483-3p-mimic groups as compared to basal; † denotes p = 0.005 on PID-10 & 0.05 on PID-13 as compared to corresponding data point in the non-targeting mimic group; and ‡ denotes p = 0.031 on PID-10, 0.022 on PID-13 as compared to corresponding data point in the vehicle group.
Change in frequency of paw withdrawal to plantar application of a von Frey filament force of 0.07 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered miR-291b-5p mimic (red symbols) or non-targeting mimic (green) or vehicle (grey symbols). * denotes p < 0.0001 from PID-3 through PID-14 in vehicle, non-targeting-mimic and miR-291b-5p-mimic groups.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 50% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally deliveredmiR-291b-5p mimic (red bars) or non-targeting mimic (green bars) or vehicle (grey bars). * denotes p < 0.0001 from PID-3 through PID-14 in vehicle, non-targeting-mimic and miR-291b-5p-mimic groups.
QRT-PCR analysis demonstrating induction of predicted miR-1a-3p target genes in lumbar DRGs in vivo following suppression of miR-1a-3p expression via inhibitor delivery intrathecally in vivo. * denotes p <:0.0001 as compared to mismatch-inhibitor group.
QRT-PCR analysis representing changes in expression of miR-1a-3p target genes in ipsilateral lumbar DRGs in vivo at day 8 in the bone metastases model. * denotes p = 0.041 as compared to sham group.
Time course of tumor-induced downregulation of Clcn3 expression in ipsilateral lumbar DRGs via QRT-PCR analysis. * denotes p = 0.002 on PID-4, 0.003 on PID-8, 0.05 on PID-10 and 0.04 on PID-15 as compared to sham group.
Time course of tumor-induced changes in miR-1a-3p expression in ipsilateral lumbar DRGs via QRT-PCR analysis. * denotes p = 0.04 on PID-4, 0.002 on PID-8, and 0.04 on PID-10 as compared to sham group.
In panels A, B, C &D statistical P value was calculated by ANOVA followed by post hoc Fischer's test, n = 3 mice per group.
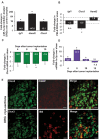
Immunohistochemical analysis of expression of Clcn3 protein in mouse DRG and colabeling with isolectin-B4-binding (IB4) non-peptidergic nociceptors and peptidergic (CGRP-positive) nociceptors. Scale bars represent 50 µm.

Binding sites for miR-1a-3p (mmu-miR-1) on the 3' untranslated region (UTR) of the mouse Clcn3 gene.
Luciferase-reporter based assay in HEK293 cells demonstrating changes in translation of the Clcn3 gene following suppression of miR-1a-3p expression via graded delivery of the specific inhibitor. *p = 0.04, 0.02 and 0.05 for 6, 4 & 2 µg/L miR-1a-3p inhibitor groups respectively as compared to control, Student's t-test, n = 3 independent experiments.
Luciferase-reporter based assay in HEK293 cells demonstrating changes in translation of the Clcn3 gene following suppression or induction of miR-1a-3p expression via graded delivery of the specific mimic. *p = 0.03, & 0.02 for 4 & 2 µg/L miR-1a-3p inhibitor groups respectively as compared to control, Student's t-test, n = 3 independent experiments.
QRT-PCR analysis demonstrating significant knockdown of Clcn3 expression in lumbar DRG following intrathecal delivery of a siRNA directed against Clcn3 or the corresponding control non-targeting siRNA, *p = 0.001 as compared to naive group, ANOVA followed by post hoc Fischer's test, n = 3 mice per group.
Change in frequency of paw withdrawal to plantar application of von Frey filament forces of 0.02 g following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally delivered siRNA directed against Clcn3 (red symbols) or the corresponding non-targeting siRNA (green symbols) or vehicle (grey symbols). * denotes p = 0.0006 on PID-11, and <0.0001 on PID-13 & 15 in the vehicle group, <0.0001 on PID-15 in the non-targeting siRNA group, 0.0037 on PID-5, and <0.0001 on PID-7 through 15 for the Clcn3-siRNA group as compared to basal, † and ‡ denotes p = 0.0202 on PID-5, 0.0011 on PID-7 and <0.0001 on PID-9, 11, 13 & 15 as compared to corresponding data point in the non-targeting siRNA or vehicle groups respectively. The square box represents the time-course of Clcn3-siRNA or non-targeting-siRNA or vehicle application.
Mechanical response thresholds calculated as von Frey filament strength required to achieve 80% withdrawal frequency following induction of tumor growth in the calcaneous bone of the heel in mice receiving intrathecally deliveredsiRNA directed against Clcn3 (red bars) or the corresponding non-targeting siRNA (green bars) or vehicle (grey bars). * denotes p = 0.0003 on PID-5 & 9, 0.0233 on PID-7, and <0.0001 on PID-11, 13 & 15 in the vehicle group, 0.0003 on PID-5, 0.002 on PID-7 and <0.0001 on PID-9, 11, 13 & 15 for the non-targeting-siRNA group and <0.0001 from PID-5 through PID-15 for the Clcn3-siRNA group as compared to basal; † denotes p = 0.0122 on PID-5, 0.0024 on PID-7, 0.0127 on PID-9, 0.05 on PID-11 as compared to corresponding data point in the non-targeting-siRNA group and ‡ denotes p = 0.0122 on PID-5, 0.0002 on PID-7, 0.0029 on PID-9, and 0.0401 on PID-11 as compared to corresponding data point in the vehicle group.
References
-
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. - PubMed
-
- Bastian I, Tam Tam S, Zhou XF, Kazenwadel J, Van der Hoek M, Michael MZ, Gibbins I, Haberberger RV. Differential expression of microRNA-1 in dorsal root ganglion neurons. Histochem Cell Biol. 2011;135:37–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

