Dynamics of DNA methylation in recent human and great ape evolution
- PMID: 24039605
- PMCID: PMC3764194
- DOI: 10.1371/journal.pgen.1003763
Dynamics of DNA methylation in recent human and great ape evolution
Abstract
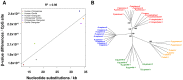
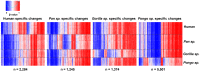
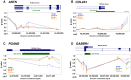
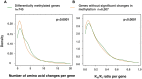
DNA methylation is an epigenetic modification involved in regulatory processes such as cell differentiation during development, X-chromosome inactivation, genomic imprinting and susceptibility to complex disease. However, the dynamics of DNA methylation changes between humans and their closest relatives are still poorly understood. We performed a comparative analysis of CpG methylation patterns between 9 humans and 23 primate samples including all species of great apes (chimpanzee, bonobo, gorilla and orangutan) using Illumina Methylation450 bead arrays. Our analysis identified ∼800 genes with significantly altered methylation patterns among the great apes, including ∼170 genes with a methylation pattern unique to human. Some of these are known to be involved in developmental and neurological features, suggesting that epigenetic changes have been frequent during recent human and primate evolution. We identified a significant positive relationship between the rate of coding variation and alterations of methylation at the promoter level, indicative of co-occurrence between evolution of protein sequence and gene regulation. In contrast, and supporting the idea that many phenotypic differences between humans and great apes are not due to amino acid differences, our analysis also identified 184 genes that are perfectly conserved at protein level between human and chimpanzee, yet show significant epigenetic differences between these two species. We conclude that epigenetic alterations are an important force during primate evolution and have been under-explored in evolutionary comparative genomics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- The Chimpanzee Sequencing AC (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 doi:10.1038/nature04072 - DOI - PubMed
-
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, et al. (2012) Insights into hominid evolution from the gorilla genome sequence. Nature 483: 169–175 doi:10.1038/nature10842 - DOI - PMC - PubMed
-
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, et al. (2012) The bonobo genome compared with the chimpanzee and human genomes. Nature 486: 527–31 doi:10.1038/nature11128 - DOI - PMC - PubMed
-
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth L V, et al. (2011) Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533 doi:10.1038/nature09687 - DOI - PMC - PubMed
-
- Cain CE, Blekhman R, Marioni JC, Gilad Y (2011) Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics 187: 1225–1234 doi:10.1534/genetics.110.126177 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

