Cell-type specific features of circular RNA expression
- PMID: 24039610
- PMCID: PMC3764148
- DOI: 10.1371/journal.pgen.1003777
Cell-type specific features of circular RNA expression
Erratum in
- PLoS Genet. 2013 Dec;9(12). doi:10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855
Abstract
Thousands of loci in the human and mouse genomes give rise to circular RNA transcripts; at many of these loci, the predominant RNA isoform is a circle. Using an improved computational approach for circular RNA identification, we found widespread circular RNA expression in Drosophila melanogaster and estimate that in humans, circular RNA may account for 1% as many molecules as poly(A) RNA. Analysis of data from the ENCODE consortium revealed that the repertoire of genes expressing circular RNA, the ratio of circular to linear transcripts for each gene, and even the pattern of splice isoforms of circular RNAs from each gene were cell-type specific. These results suggest that biogenesis of circular RNA is an integral, conserved, and regulated feature of the gene expression program.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

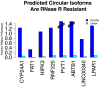

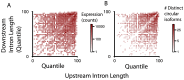




References
-
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, et al. (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73: 1019–1030. - PubMed
-
- Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O'Dell KM, et al. (2006) Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. The Journal of heredity 97: 253–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

