Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15
- PMID: 24039739
- PMCID: PMC3765158
- DOI: 10.1371/journal.pone.0072114
Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15
Abstract
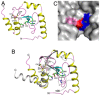
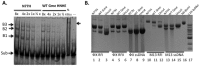
The crystal structure of a putative HNH endonuclease, Gmet_0936 protein from Geobacter metallireducens GS-15, has been determined at 2.6 Å resolution using single-wavelength anomalous dispersion method. The structure contains a two-stranded anti-parallel β-sheet that are surrounded by two helices on each face, and reveals a Zn ion bound in each monomer, coordinated by residues Cys38, Cys41, Cys73, and Cys76, which likely plays an important structural role in stabilizing the overall conformation. Structural homologs of Gmet_0936 include Hpy99I endonuclease, phage T4 endonuclease VII, and other HNH endonucleases, with these enzymes sharing 15-20% amino acid sequence identity. An overlay of Gmet_0936 and Hpy99I structures shows that most of the secondary structure elements, catalytic residues as well as the zinc binding site (zinc ribbon) are conserved. However, Gmet_0936 lacks the N-terminal domain of Hpy99I, which mediates DNA binding as well as dimerization. Purified Gmet_0936 forms dimers in solution and a dimer of the protein is observed in the crystal, but with a different mode of dimerization as compared to Hpy99I. Gmet_0936 and its N77H variant show a weak DNA binding activity in a DNA mobility shift assay and a weak Mn²⁺-dependent nicking activity on supercoiled plasmids in low pH buffers. The preferred substrate appears to be acid and heat-treated DNA with AP sites, suggesting Gmet_0936 may be a DNA repair enzyme.
Conflict of interest statement
Figures



Similar articles
-
Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes.J Biol Chem. 2013 Mar 22;288(12):8445-8455. doi: 10.1074/jbc.M112.422774. Epub 2013 Jan 25. J Biol Chem. 2013. PMID: 23355472 Free PMC article.
-
Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.Biochimie. 2016 Sep-Oct;128-129:20-33. doi: 10.1016/j.biochi.2016.06.011. Epub 2016 Jun 22. Biochimie. 2016. PMID: 27343627
-
Characterization of biochemical properties of an apurinic/apyrimidinic endonuclease from Helicobacter pylori.PLoS One. 2018 Aug 15;13(8):e0202232. doi: 10.1371/journal.pone.0202232. eCollection 2018. PLoS One. 2018. PMID: 30110394 Free PMC article.
-
Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
-
The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once.DNA Repair (Amst). 2014 Jul;19:95-107. doi: 10.1016/j.dnarep.2014.03.022. Epub 2014 Apr 19. DNA Repair (Amst). 2014. PMID: 24754999 Free PMC article. Review.
Cited by
-
Phylogenomics and sequence-structure-function relationships in the GmrSD family of Type IV restriction enzymes.BMC Bioinformatics. 2015 Oct 23;16:336. doi: 10.1186/s12859-015-0773-z. BMC Bioinformatics. 2015. PMID: 26493560 Free PMC article.
-
Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease.Sci Rep. 2017 Feb 13;7:42542. doi: 10.1038/srep42542. Sci Rep. 2017. PMID: 28211904 Free PMC article.
-
Structural insights into the function of ZRANB3 in replication stress response.Nat Commun. 2017 Jun 16;8:15847. doi: 10.1038/ncomms15847. Nat Commun. 2017. PMID: 28621305 Free PMC article.
-
Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases.Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):6016-21. doi: 10.1073/pnas.1320538111. Epub 2014 Apr 7. Proc Natl Acad Sci U S A. 2014. PMID: 24711396 Free PMC article.
-
Engineering nicking enzymes that preferentially nick 5-methylcytosine-modified DNA.Nucleic Acids Res. 2014 May;42(9):e77. doi: 10.1093/nar/gku192. Epub 2014 Mar 7. Nucleic Acids Res. 2014. PMID: 24609382 Free PMC article.
References
-
- Liu C, Gorby YA, Zachara JM, Fredrickson JK, Brown CF (2002) Reduction kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in cultures of dissimilatory metal-reducing bacteria. Biotechnology and bioengineering 80: 637–649. - PubMed
-
- Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, et al. (1993) Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Archives of microbiology 159: 336–344. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

