Continuous influenza virus production in cell culture shows a periodic accumulation of defective interfering particles
- PMID: 24039749
- PMCID: PMC3764112
- DOI: 10.1371/journal.pone.0072288
Continuous influenza virus production in cell culture shows a periodic accumulation of defective interfering particles
Abstract
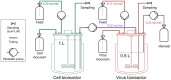
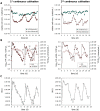
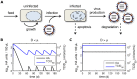
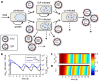
Influenza viruses are a major public health burden during seasonal epidemics and a continuous threat due to their potential to cause pandemics. Annual vaccination provides the best protection against the contagious respiratory illness caused by influenza viruses. However, the current production capacities for influenza vaccines are insufficient to meet the increasing demands. We explored the possibility to establish a continuous production process for influenza viruses using the duck-derived suspension cell line AGE1.CR. A two-stage bioreactor setup was designed in which cells were cultivated in a first stirred tank reactor where an almost constant cell concentration was maintained. Cells were then constantly fed to a second bioreactor where virus infection and replication took place. Using this two-stage reactor system, it was possible to continuously produce influenza viruses. Surprisingly, virus titers showed a periodic increase and decrease during the run-time of 17 days. These titer fluctuations were caused by the presence of defective interfering particles (DIPs), which we detected by PCR. Mathematical modeling confirmed this observation showing that constant virus titers can only emerge in the absence of DIPs. Even with very low amounts of DIPs in the seed virus and very low rates for de novo DIP generation, defective viruses rapidly accumulate and, therefore, represent a serious challenge for continuous vaccine production. Yet, the continuous replication of influenza virus using a two-stage bioreactor setup is a novel tool to study aspects of viral evolution and the impact of DIPs.
Conflict of interest statement
Figures

References
-
- Zuccotti GV, Fabiano V (2011) Strategies for preventing influenza: future perspectives in influenza vaccine technology. Expert Opinion on Biological Therapy 11: 1–4. - PubMed
-
- Audsley JM, Tannock GA (2008) Cell-based influenza vaccines: progress to date. Drugs 68: 1483–1491. - PubMed
-
- Genzel Y, Reichl U (2009) Continuous cell lines as a production system for influenza vaccines. Expert Review of Vaccines 8: 1681–1692. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

