The stack: a new bacterial structure analyzed in the Antarctic bacterium Pseudomonas deceptionensis M1(T) by transmission electron microscopy and tomography
- PMID: 24039905
- PMCID: PMC3767748
- DOI: 10.1371/journal.pone.0073297
The stack: a new bacterial structure analyzed in the Antarctic bacterium Pseudomonas deceptionensis M1(T) by transmission electron microscopy and tomography
Abstract
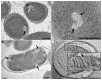
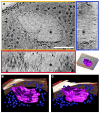
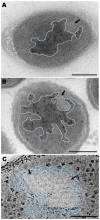
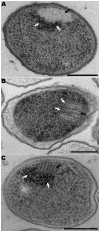
In recent years, improvements in transmission electron microscopy (TEM) techniques and the use of tomography have provided a more accurate view of the complexity of the ultrastructure of prokaryotic cells. Cryoimmobilization of specimens by rapid cooling followed by freeze substitution (FS) and sectioning, freeze fracture (FF) and observation of replica, or cryoelectron microscopy of vitreous sections (CEMOVIS) now allow visualization of biological samples close to their native state, enabling us to refine our knowledge of already known bacterial structures and to discover new ones. Application of these techniques to the new Antarctic cold-adapted bacterium Pseudomonasdeceptionensis M1(T) has demonstrated the existence of a previously undescribed cytoplasmic structure that does not correspond to known bacterial inclusion bodies or membranous formations. This structure, which we term a "stack", was mainly visualized in slow growing cultures of P. deceptionensis M1(T) and can be described as a set of stacked membranous discs usually arranged perpendicularly to the cell membrane, but not continuous with it, and found in variable number in different locations within the cell. Regardless of their position, stacks were mostly observed very close to DNA fibers. Stacks are not exclusive to P. deceptionensis M1(T) and were also visualized in slow-growing cultures of other bacteria. This new structure deserves further study using cryoelectron tomography to refine its configuration and to establish whether its function could be related to chromosome dynamics.
Conflict of interest statement
Figures



References
-
- Shively JM, Ball F, Brown DH, Saunders RE (1973) Functional Organelles in Prokaryotes: Polyhedral Inclusions (Carboxysomes) of Thiobacillus neapolitanus. Science 182: 584–586. doi:10.1126/science.182.4112.584. PubMed: 4355679. - DOI - PubMed
-
- Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA et al. (2008) Biochemical and Structural Insights into Bacterial Organelle Form and Biogenesis. J Biol Chem 283: 14366–14375. doi:10.1074/jbc.M709214200. PubMed: 18332146. - DOI - PubMed
-
- O’Connell JD, Zhao A, Ellington AD, Marcotte EM (2012) Dynamic Reorganization of Metabolic Enzymes into Intracellular Bodies. Annu Rev Cell Dev Biol 28: 89–111. doi:10.1146/annurev-cellbio-101011-155841. PubMed: 23057741. - DOI - PMC - PubMed
-
- Shively JM (1974) Inclusion bodies of prokaryotes. Annu Rev Microbiol 28: 167–187. doi:10.1146/annurev.mi.28.100174.001123. PubMed: 4372937. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

