Multivalent engagement of TFIID to nucleosomes
- PMID: 24039962
- PMCID: PMC3770614
- DOI: 10.1371/journal.pone.0073495
Multivalent engagement of TFIID to nucleosomes
Abstract
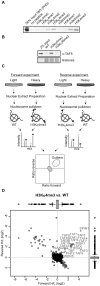
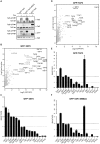
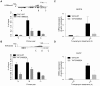
The process of eukaryotic transcription initiation involves the assembly of basal transcription factor complexes on the gene promoter. The recruitment of TFIID is an early and important step in this process. Gene promoters contain distinct DNA sequence elements and are marked by the presence of post-translationally modified nucleosomes. The contributions of these individual features for TFIID recruitment remain to be elucidated. Here, we use immobilized reconstituted promoter nucleosomes, conventional biochemistry and quantitative mass spectrometry to investigate the influence of distinct histone modifications and functional DNA-elements on the binding of TFIID. Our data reveal synergistic effects of H3K4me3, H3K14ac and a TATA box sequence on TFIID binding in vitro. Stoichiometry analyses of affinity purified human TFIID identified the presence of a stable dimeric core. Several peripheral TAFs, including those interacting with distinct promoter features, are substoichiometric yet present in substantial amounts. Finally, we find that the TAF3 subunit of TFIID binds to poised promoters in an H3K4me3-dependent manner. Moreover, the PHD-finger of TAF3 is important for rapid induction of target genes. Thus, fine-tuning of TFIID engagement on promoters is driven by synergistic contacts with both DNA-elements and histone modifications, eventually resulting in a high affinity interaction and activation of transcription.
Conflict of interest statement
Figures

References
-
- Buratowski S, Hahn S, Guarente L, Sharp PA (1989) Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561. - PubMed
-
- Thomas MC, Chiang CM (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178. - PubMed
-
- Gangloff YG, Romier C, Thuault S, Werten S, Davidson I (2001) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 26: 250–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

