Chromatin changes in dicer-deficient mouse embryonic stem cells in response to retinoic acid induced differentiation
- PMID: 24040281
- PMCID: PMC3767645
- DOI: 10.1371/journal.pone.0074556
Chromatin changes in dicer-deficient mouse embryonic stem cells in response to retinoic acid induced differentiation
Abstract
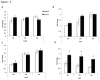
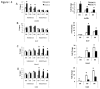
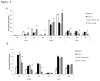
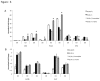
Loss of Dicer, an enzyme critical for microRNA biogenesis, results in lethality due to a block in mouse embryonic stem cell (mES) differentiation. Using ChIP-Seq we found increased H3K9me2 at over 900 CpG islands in the Dicer(-/-)ES epigenome. Gene ontology analysis revealed that promoters of chromatin regulators to be among the most impacted by increased CpG island H3K9me2 in ES (Dicer(-/-)). We therefore, extended the study to include H3K4me3 and H3K27me3 marks for selected genes. We found that the ES (Dicer(-/-)) mutant epigenome was characterized by a shift in the overall balance between transcriptionally favorable (H3K4me3) and unfavorable (H3K27me3) marks at key genes regulating ES cell differentiation. Pluripotency genes Oct4, Sox2 and Nanog were not impacted in relation to patterns of H3K27me3 and H3K4me3 and showed no changes in the rates of transcript down-regulation in response to RA. The most striking changes were observed in regards to genes regulating differentiation and the transition from self-renewal to differentiation. An increase in H3K4me3 at the promoter of Lin28b was associated with the down-regulation of this gene at a lower rate in Dicer(-/-)ES as compared to wild type ES. An increase in H3K27me3 in the promoters of differentiation genes Hoxa1 and Cdx2 in Dicer(-/-)ES cells was coincident with an inability to up-regulate these genes at the same rate as ES upon retinoic acid (RA)-induced differentiation. We found that siRNAs Ezh2 and post-transcriptional silencing of Ezh2 by let-7 g rescued this effect suggesting that Ezh2 up-regulation is in part responsible for increased H3K27me3 and decreased rates of up-regulation of differentiation genes in Dicer(-/-)ES.
Conflict of interest statement
Figures


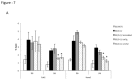
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. doi:10.1038/35053110. PubMed: 11201747. - DOI - PubMed
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. doi:10.1016/S0092-8674(04)00045-5. PubMed: 14744438. - DOI - PubMed
-
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS et al. (2006) Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 124: 355-366. doi:10.1016/j.cell.2005.11.034. PubMed: 16439209. - DOI - PubMed
-
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN et al. (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311: 195-198. doi:10.1126/science.1121638. PubMed: 16410517. - DOI - PubMed
-
- Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7: 653-662. doi:10.1016/j.devcel.2004.10.003. PubMed: 15525527. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

