Synergistic re-activation of epigenetically silenced genes by combinatorial inhibition of DNMTs and LSD1 in cancer cells
- PMID: 24040395
- PMCID: PMC3765366
- DOI: 10.1371/journal.pone.0075136
Synergistic re-activation of epigenetically silenced genes by combinatorial inhibition of DNMTs and LSD1 in cancer cells
Abstract
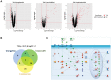
Epigenetic gene silencing, mediated by aberrant promoter DNA hypermethylation and repressive histone modifications, is a hallmark of cancer. Although heritable, the dynamic nature and potential reversibility through pharmacological interventions make such aberrations attractive targets. Since cancers contain multiple epigenetic abnormalities, combining therapies that target different defects could potentially enhance their individual efficacies. 5-Aza-2'-deoxycytidine (5-Aza-CdR), FDA-approved drug for the treatment of myelodysplastic syndrome, can inhibit DNA methyltransferases (DNMTs) upon incorporation into the DNA of dividing cells, resulting in global demethylation. More recently, the first histone demethylase, lysine specific demethylase 1 (LSD1), which demethylates both histone and non-histone substrates, has become a new target for epigenetic therapy. Using, clorgyline, an LSD1 inhibitor (LSD1i) to treat cancer cell lines, we show that clorgyline employs two mechanisms of action depending on the cell type: it can either induce global DNA demethylation or inhibit LSD1-driven H3K4me2 and H3K4me1 demethylation to establish an active chromatin configuration. We also investigate the therapeutic efficacy of combining 5-Aza-CdR with clorgyline and determine that this combinatorial treatment has synergistic effects on reactivating aberrantly silenced genes by enriching H3K4me2 and H3K4me1. Many of the reactivated genes are categorized as cancer testis antigens or belong to the interferon-signaling pathway, suggesting potential implications for immunotherapy. Together, our results demonstrate that combinatorial treatment consisting of a DNMT inhibitor (DNMTi) and an LSD1i have enhanced therapeutic values and could improve the efficacy of epigenetic therapy.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

