Bias problems in culture-independent analysis of environmental bacterial communities: a representative study on hydrocarbonoclastic bacteria
- PMID: 24040582
- PMCID: PMC3769543
- DOI: 10.1186/2193-1801-2-369
Bias problems in culture-independent analysis of environmental bacterial communities: a representative study on hydrocarbonoclastic bacteria
Abstract
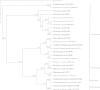
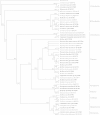
Culture-dependent methods for bacterial community analysis are currently considered obsolete; therefore, molecular techniques are usually used instead. The results of the current study on hydrocarbonoclastic bacteria in various oily habitats in Kuwait showed however, that the bacterial identities varied dramatically according to the analytical approach used. For six desert and six seawater samples used in this study, the culture-independent and culture-dependent techniques each led to a unique bacterial composition. Problems related to the culture-dependent technique are well known. The results of the current study highlighted bias problems other than those already recorded in the literature for the molecular approaches. Thus, for example, in contrast to the culture-dependent technique, the primers used in the molecular approach preferentially amplified the 16S rDNAs of hydrocarbonoclastic bacteria in total genomic DNAs of all the studied environmental samples, and in addition, failed to reveal in any environmental sample members of the Actinobacteria. The primers used in the molecular approach also amplified certain "pure" 16S rDNAs, but failed to do so when these DNAs were in mixture. In view of these results, it is recommended that the two analytical approaches should be used simultaneously because their combined results would reflect the bacterial community composition more precisely than either of them can do alone.
Figures



References
-
- Al-Bader D, Kansour M, Rayan R, Radwan SS. Biofilm comprising 354 phototrophic, diazotrophic, and hydrocarbon-utilizing bacteria: a promising 355 consortium in the bioremediation of aquatic hydrocarbon pollutants. Environ Sci Pollut Res (2013) 2012;20:3252–3262. doi: 10.1007/s11356-012-1251-z. - DOI - PubMed
-
- Al-Sarawi HA, Mahmoud HM, Radwan SS. Pyruvate-utilizing bacteria as contributors to the food web in the Arabian Gulf. Mar Biol. 2008;154:337–381. doi: 10.1007/s00227-008-0937-8. - DOI
-
- Arun K, Ashok M, Rajesh S. Crude oil PAH constitution, degradation pathway and associated bioremediation microflora: an overview. Int J Environ Sci. 2011;1:1420–1439.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

