Space-efficient and exact de Bruijn graph representation based on a Bloom filter
- PMID: 24040893
- PMCID: PMC3848682
- DOI: 10.1186/1748-7188-8-22
Space-efficient and exact de Bruijn graph representation based on a Bloom filter
Abstract
Background: The de Bruijn graph data structure is widely used in next-generation sequencing (NGS). Many programs, e.g. de novo assemblers, rely on in-memory representation of this graph. However, current techniques for representing the de Bruijn graph of a human genome require a large amount of memory (≥30 GB).
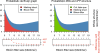
Results: We propose a new encoding of the de Bruijn graph, which occupies an order of magnitude less space than current representations. The encoding is based on a Bloom filter, with an additional structure to remove critical false positives.
Conclusions: An assembly software implementing this structure, Minia, performed a complete de novo assembly of human genome short reads using 5.7 GB of memory in 23 hours.
Figures


References
-
- Grabherr MG. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;8(7):644–652. doi: 10.1038/nbt.1883. [ http://dx.doi.org/10.1038/nbt.1883] - DOI - DOI - PMC - PubMed
-
- Peterlongo P, Schnel N, Pisanti N, Sagot MF, Lacroix V. String Processing and Information Retrieval. Berlin, Heidelberg: Springer; 2010. Identifying SNPs without a reference genome by comparing raw reads; pp. 147–158.
LinkOut - more resources
Full Text Sources
Other Literature Sources

