Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection
- PMID: 24041034
- PMCID: PMC3849974
- DOI: 10.1186/1742-4690-10-98
Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection
Abstract
Background: Human immunodeficiency virus type 1 (HIV-1) subtype C (C-HIV) is spreading rapidly and is now responsible for >50% of HIV-1 infections worldwide, and >95% of infections in southern Africa and central Asia. These regions are burdened with the overwhelming majority of HIV-1 infections, yet we know very little about the pathogenesis of C-HIV. In addition to CCR5 and CXCR4, the HIV-1 envelope glycoproteins (Env) may engage a variety of alternative coreceptors for entry into transfected cells. Whilst alternative coreceptors do not appear to have a broad role in mediating the entry of HIV-1 into primary cells, characterizing patterns of alternative coreceptor usage in vitro can provide valuable insights into mechanisms of Env-coreceptor engagement that may be important for HIV-1 pathogenesis.
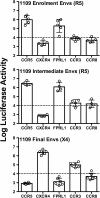
Results: Here, we characterized the ability of luciferase reporter viruses pseudotyped with HIV-1 Envs (n = 300) cloned sequentially from plasma of 21 antiretroviral therapy (ART)-naïve subjects experiencing progression from chronic to advanced C-HIV infection over an approximately 3-year period, who either exclusively maintained CCR5-using (R5) variants (n = 20 subjects) or who experienced a coreceptor switch to CXCR4-using (X4) variants (n = 1 subject), to utilize alternative coreceptors for entry. At a population level, CCR5 usage by R5 C-HIV Envs was strongly linked to usage of FPRL1, CCR3 and CCR8 as alternative coreceptors, with the linkages to FPRL1 and CCR3 usage becoming statistically more robust as infection progressed from chronic to advanced stages of disease. In contrast, acquisition of an X4 Env phenotype at advanced infection was accompanied by a dramatic loss of FPRL1 usage. Env mutagenesis studies confirmed a direct link between CCR5 and FPRL1 usage, and showed that the V3 loop crown, but not other V3 determinants of CCR5-specificity, was the principal Env determinant governing the ability of R5 C-HIV Envs from one particular subject to engage FPRL1.
Conclusions: Our results suggest that, in the absence of coreceptor switching, the ability of R5 C-HIV viruses to engage certain alternative coreceptors in vitro, in particular FPRL1, may reflect an altered use of CCR5 that is selected for during progressive C-HIV infection, and which may contribute to C-HIV pathogenicity.
Figures



Similar articles
-
Virus entry via the alternative coreceptors CCR3 and FPRL1 differs by human immunodeficiency virus type 1 subtype.J Virol. 2009 Sep;83(17):8353-63. doi: 10.1128/JVI.00780-09. Epub 2009 Jun 24. J Virol. 2009. PMID: 19553323 Free PMC article.
-
Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection.Infect Genet Evol. 2010 Apr;10(3):356-64. doi: 10.1016/j.meegid.2009.05.003. Epub 2009 May 14. Infect Genet Evol. 2010. PMID: 19446658
-
High-Sequence Diversity and Rapid Virus Turnover Contribute to Higher Rates of Coreceptor Switching in Treatment-Experienced Subjects with HIV-1 Viremia.AIDS Res Hum Retroviruses. 2017 Mar;33(3):234-245. doi: 10.1089/AID.2016.0153. Epub 2016 Oct 12. AIDS Res Hum Retroviruses. 2017. PMID: 27604829 Free PMC article.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
Cited by
-
Differences in coreceptor specificity contribute to alternative tropism of HIV-1 subtype C for CD4(+) T-cell subsets, including stem cell memory T-cells.Retrovirology. 2014 Nov 12;11:97. doi: 10.1186/s12977-014-0097-5. Retrovirology. 2014. PMID: 25387392 Free PMC article.
-
Distinct HIV-1 entry phenotypes are associated with transmission, subtype specificity, and resistance to broadly neutralizing antibodies.Retrovirology. 2014 Jun 23;11:48. doi: 10.1186/1742-4690-11-48. Retrovirology. 2014. PMID: 24957778 Free PMC article.
-
Frequency and Env determinants of HIV-1 subtype C strains from antiretroviral therapy-naive subjects that display incomplete inhibition by maraviroc.Retrovirology. 2016 Nov 3;13(1):74. doi: 10.1186/s12977-016-0309-2. Retrovirology. 2016. PMID: 27809912 Free PMC article.
-
HIV Entry and Its Inhibition by Bifunctional Antiviral Proteins.Mol Ther Nucleic Acids. 2018 Dec 7;13:347-364. doi: 10.1016/j.omtn.2018.09.003. Epub 2018 Sep 11. Mol Ther Nucleic Acids. 2018. PMID: 30340139 Free PMC article. Review.
-
Unique Phenotypic Characteristics of Recently Transmitted HIV-1 Subtype C Envelope Glycoprotein gp120: Use of CXCR6 Coreceptor by Transmitted Founder Viruses.J Virol. 2018 Apr 13;92(9):e00063-18. doi: 10.1128/JVI.00063-18. Print 2018 May 1. J Virol. 2018. PMID: 29491151 Free PMC article.
References
-
- Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N. et al.Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186(3):405–411. doi: 10.1084/jem.186.3.405. - DOI - PMC - PubMed
-
- Gorry PR, Dunfee RL, Mefford ME, Kunstman K, Morgan T, Moore JP, Mascola JR, Agopian K, Holm GH, Mehle A. et al.Changes in the V3 region of gp120 contribute to unusually broad coreceptor usage of an HIV-1 isolate from a CCR5 Delta32 heterozygote. Virology. 2007;362(1):163–178. doi: 10.1016/j.virol.2006.11.025. - DOI - PMC - PubMed
-
- McKnight A, Dittmar MT, Moniz-Periera J, Ariyoshi K, Reeves JD, Hibbitts S, Whitby D, Aarons E, Proudfoot AE, Whittle H. et al.A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72(5):4065–4071. - PMC - PubMed
-
- Reeves JD, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira JM, Moniz-Pereira J, Clapham PR. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73(9):7795–7804. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

