Membrane protein structure determination in membrana
- PMID: 24041243
- PMCID: PMC3970975
- DOI: 10.1021/ar400041a
Membrane protein structure determination in membrana
Abstract
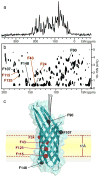
The two principal components of biological membranes, the lipid bilayer and the proteins integrated within it, have coevolved for specific functions that mediate the interactions of cells with their environment. Molecular structures can provide very significant insights about protein function. In the case of membrane proteins, the physical and chemical properties of lipids and proteins are highly interdependent; therefore structure determination should include the membrane environment. Considering the membrane alongside the protein eliminates the possibility that crystal contacts or detergent molecules could distort protein structure, dynamics, and function and enables ligand binding studies to be performed in a natural setting. Solid-state NMR spectroscopy is compatible with three-dimensional structure determination of membrane proteins in phospholipid bilayer membranes under physiological conditions and has played an important role in elucidating the physical and chemical properties of biological membranes, providing key information about the structure and dynamics of the phospholipid components. Recently, developments in the recombinant expression of membrane proteins, sample preparation, pulse sequences for high-resolution spectroscopy, radio frequency probes, high-field magnets, and computational methods have enabled a number of membrane protein structures to be determined in lipid bilayer membranes. In this Account, we illustrate solid-state NMR methods with examples from two bacterial outer membrane proteins (OmpX and Ail) that form integral membrane β-barrels. The ability to measure orientation-dependent frequencies in the solid-state NMR spectra of membrane-embedded proteins provides the foundation for a powerful approach to structure determination based primarily on orientation restraints. Orientation restraints are particularly useful for NMR structural studies of membrane proteins because they provide information about both three-dimensional structure and the orientation of the protein within the membrane. When combined with dihedral angle restraints derived from analysis of isotropic chemical shifts, molecular fragment replacement, and de novo structure prediction, orientation restraints can yield high-quality three-dimensional structures with few or no distance restraints. Using complementary solid-state NMR methods based on oriented sample (OS) and magic angle spinning (MAS) approaches, one can resolve and assign multiple peaks through the use of (15)N/(13)C labeled samples and measure precise restraints to determine structures.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- McLaughlin AC, Cullis PR, Hemminga MA, Hoult DI, Radda GK, Ritchie GA, Seeley PJ, Richards RE. Application of 31P NMR to model and biological membrane systems. FEBS Lett. 1975;57:213–218. - PubMed
-
- Griffin RG. Letter: Observation of the effect of water on the 31P nuclear magnetic resonance spectra of dipalmitoyllecithin. J Am Chem Soc. 1976;98:851–853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

