Methamphetamine alters reference gene expression in nigra and striatum of adult rat brain
- PMID: 24042092
- PMCID: PMC3918345
- DOI: 10.1016/j.neuro.2013.08.009
Methamphetamine alters reference gene expression in nigra and striatum of adult rat brain
Abstract
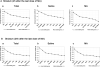
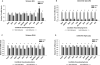
The nigrostriatal dopaminergic system is a major lesion target for methamphetamine (MA), one of the most addictive and neurotoxic drugs of abuse. High doses of MA alter the expression of a large number of genes. Reference genes (RGs) are considered relatively stable and are often used as standards for quantitative real-time PCR (qRT-PCR) reactions. The purpose of this study was to determine whether MA altered the expression of RGs and to identify the appropriate RGs for gene expression studies in animals receiving MA. Adult male Sprague-Dawley rats were treated with high doses of MA or saline. Striatum and substantia nigra were harvested at 2h or 24h after MA administration. The expression and stability of 10 commonly used RGs were examined using qRT-PCR and then evaluated by geNorm and Normfinder. We found that MA altered the expression of selected RGs. These candidate RGs presented differential stability in the striatum and in substantia nigra at both 2h and 24h after MA injection. Selection of an unstable RG as a standard altered the significance of tyrosine hydroxylase (TH) mRNA expression after MA administration. In conclusion, our data show that MA site- and time-dependently altered the expression of RGs in nigrostriatal dopaminergic system. These temporal and spatial factors should be considered when selecting appropriate RGs for interpreting the expression of target genes in animals receiving MA.
Keywords: Housekeeping genes; Methamphetamine; Nigra; Real-time PCR; Reference genes; Striatum.
Published by Elsevier B.V.
Figures


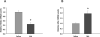
Similar articles
-
9-Cis retinoic acid protects against methamphetamine-induced neurotoxicity in nigrostriatal dopamine neurons.Neurotox Res. 2014 Apr;25(3):248-61. doi: 10.1007/s12640-013-9413-4. Epub 2013 Jul 25. Neurotox Res. 2014. PMID: 23884514 Free PMC article.
-
Escalating Methamphetamine Regimen Induces Compensatory Mechanisms, Mitochondrial Biogenesis, and GDNF Expression, in Substantia Nigra.J Cell Biochem. 2017 Jun;118(6):1369-1378. doi: 10.1002/jcb.25795. Epub 2017 Jan 5. J Cell Biochem. 2017. PMID: 27862224
-
Increasing methamphetamine doses inhibit glycogen synthase kinase 3β activity by stimulating the insulin signaling pathway in substantia nigra.J Cell Biochem. 2018 Nov;119(10):8522-8530. doi: 10.1002/jcb.27082. Epub 2018 Jul 16. J Cell Biochem. 2018. PMID: 30011098
-
Epothilone D prevents binge methamphetamine-mediated loss of striatal dopaminergic markers.J Neurochem. 2016 Feb;136(3):510-25. doi: 10.1111/jnc.13391. Epub 2015 Dec 10. J Neurochem. 2016. PMID: 26465779 Free PMC article.
-
Neonatal methamphetamine in the rat: evidence for gender-specific differences upon tyrosine hydroxylase enzyme in the dopaminergic nigrostriatal system.Ann N Y Acad Sci. 2000 Sep;914:431-8. doi: 10.1111/j.1749-6632.2000.tb05217.x. Ann N Y Acad Sci. 2000. PMID: 11085342
Cited by
-
Matrix Metalloproteinase-9 Signaling Regulates Colon Barrier Integrity in Models of HIV Infection.J Neuroimmune Pharmacol. 2024 Nov 5;19(1):57. doi: 10.1007/s11481-024-10158-2. J Neuroimmune Pharmacol. 2024. PMID: 39499375
-
Identification of optimal endogenous reference RNAs for RT-qPCR normalization in hindgut of rat models with anorectal malformations.PeerJ. 2019 Apr 23;7:e6829. doi: 10.7717/peerj.6829. eCollection 2019. PeerJ. 2019. PMID: 31065464 Free PMC article.
-
Selection and Validation of Reference Genes for RT-PCR Expression Analysis of Candidate Genes Involved in Morphine-Induced Conditioned Place Preference Mice.J Mol Neurosci. 2018 Dec;66(4):587-594. doi: 10.1007/s12031-018-1198-8. Epub 2018 Nov 1. J Mol Neurosci. 2018. PMID: 30386959
-
With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies.PLoS One. 2015 Nov 10;10(11):e0141853. doi: 10.1371/journal.pone.0141853. eCollection 2015. PLoS One. 2015. PMID: 26555275 Free PMC article.
-
Validation of reference genes for quantitative real-time PCR in valproic acid rat models of autism.Mol Biol Rep. 2016 Aug;43(8):837-47. doi: 10.1007/s11033-016-4015-x. Epub 2016 Jun 10. Mol Biol Rep. 2016. PMID: 27287459
References
-
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. - PubMed
-
- Bonefeld BE, Elfving B, Wegener G. Reference genes for normalization: a study of rat brain tissue. Synapse. 2008;62:302–9. - PubMed
-
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

