Hydrogenosomes in the diplomonad Spironucleus salmonicida
- PMID: 24042146
- PMCID: PMC3778541
- DOI: 10.1038/ncomms3493
Hydrogenosomes in the diplomonad Spironucleus salmonicida
Abstract
Acquisition of the mitochondrion is a key event in the evolution of the eukaryotic cell, but diversification of the organelle has occurred during eukaryotic evolution. One example of such mitochondria-related organelles (MROs) are hydrogenosomes, which produce ATP by substrate-level phosphorylation with hydrogen as a byproduct. The diplomonad parasite Giardia intestinalis harbours mitosomes, another type of MRO. Here we identify MROs in the salmon parasite Spironucleus salmonicida with similar protein import and Fe-S cluster assembly machineries as in Giardia mitosomes. We find that hydrogen production is prevalent in the diplomonad genus Spironucleus, and that S. salmonicida MROs contain enzymes characteristic of hydrogenosomes. Evolutionary analyses of known hydrogenosomal components indicate their presence in the diplomonad ancestor, and subsequent loss in Giardia. Our results suggest that hydrogenosomes are metabolic adaptations predating the split between parabasalids and diplomonads, which is deeper than the split between animals and fungi in the eukaryotic tree.
Figures

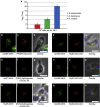

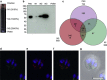
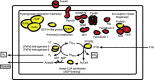
References
-
- Embley T. M. & Martin W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006). - PubMed
-
- Emelyanov V. V. & Goldberg A. V. Fermentation enzymes of Giardia intestinalis, pyruvate:ferredoxin oxidoreductase and hydrogenase, do not localize to its mitosomes. Microbiol. 157, 1602–1611 (2011). - PubMed
-
- Lloyd D., Ralphs J. R. & Harris J. C. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiol. 148, 727–733 (2002). - PubMed
-
- Tovar J. et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426, 172–176 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

