Epigenetic and genetic features of 24 colon cancer cell lines
- PMID: 24042735
- PMCID: PMC3816225
- DOI: 10.1038/oncsis.2013.35
Epigenetic and genetic features of 24 colon cancer cell lines
Abstract
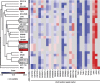
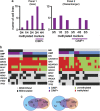
Cell lines are invaluable biomedical research tools, and recent literature has emphasized the importance of genotype authentication and characterization. In the present study, 24 out of 27 cell line identities were confirmed by short tandem repeat profiling. The molecular phenotypes of the 24 colon cancer cell lines were examined, and microsatellite instability (MSI) and CpG island methylator phenotype (CIMP) were determined, using the Bethesda panel mononucleotide repeat loci and two epimarker panels, respectively. Furthermore, the BRAF, KRAS and PIK3CA oncogenes were analyzed for mutations in known hotspots, while the entire coding sequences of the PTEN and TP53 tumor suppressors were investigated. Nine cell lines showed MSI. Thirteen and nine cell lines were found to be CIMP positive, using the Issa panel and the Weisenberger et al. panel, respectively. The latter was found to be superior for CIMP classification of colon cancer cell lines. Seventeen cell lines harbored disrupting TP53 mutations. Altogether, 20/24 cell lines had the mitogen-activated protein kinase pathway activating mutually exclusive KRAS or BRAF mutations. PIK3CA and PTEN mutations leading to hyperactivation of the phosphoinositide 3-kinase/AKT pathway were observed in 13/24 cell lines. Interestingly, in four cell lines there were no mutations in neither BRAF, KRAS, PIK3CA nor in PTEN. In conclusion, this study presents molecular features of a large number of colon cancer cell lines to aid the selection of suitable in vitro models for descriptive and functional research.
Figures

References
-
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

