SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation
- PMID: 24043310
- PMCID: PMC3838178
- DOI: 10.1128/MCB.00087-13
SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation
Abstract
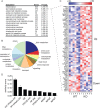
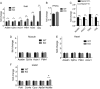
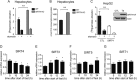
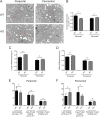
Sirtuins are a family of protein deacetylases, deacylases, and ADP-ribosyltransferases that regulate life span, control the onset of numerous age-associated diseases, and mediate metabolic homeostasis. We have uncovered a novel role for the mitochondrial sirtuin SIRT4 in the regulation of hepatic lipid metabolism during changes in nutrient availability. We show that SIRT4 levels decrease in the liver during fasting and that SIRT4 null mice display increased expression of hepatic peroxisome proliferator-activated receptor α (PPARα) target genes associated with fatty acid catabolism. Accordingly, primary hepatocytes from SIRT4 knockout (KO) mice exhibit higher rates of fatty acid oxidation than wild-type hepatocytes, and SIRT4 overexpression decreases fatty acid oxidation rates. The enhanced fatty acid oxidation observed in SIRT4 KO hepatocytes requires functional SIRT1, demonstrating a clear cross talk between mitochondrial and nuclear sirtuins. Thus, SIRT4 is a new component of mitochondrial signaling in the liver and functions as an important regulator of lipid metabolism.
Figures

Similar articles
-
SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells.J Biol Chem. 2010 Oct 15;285(42):31995-2002. doi: 10.1074/jbc.M110.124164. Epub 2010 Aug 4. J Biol Chem. 2010. PMID: 20685656 Free PMC article.
-
CDKN2A/p16INK4a suppresses hepatic fatty acid oxidation through the AMPKα2-SIRT1-PPARα signaling pathway.J Biol Chem. 2020 Dec 11;295(50):17310-17322. doi: 10.1074/jbc.RA120.012543. Epub 2020 Oct 9. J Biol Chem. 2020. PMID: 33037071 Free PMC article.
-
Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation.Cell Metab. 2009 Apr;9(4):327-38. doi: 10.1016/j.cmet.2009.02.006. Cell Metab. 2009. PMID: 19356714 Free PMC article.
-
The Role of Mitochondrial Sirtuins (SIRT3, SIRT4 and SIRT5) in Renal Cell Metabolism: Implication for Kidney Diseases.Int J Mol Sci. 2024 Jun 25;25(13):6936. doi: 10.3390/ijms25136936. Int J Mol Sci. 2024. PMID: 39000044 Free PMC article. Review.
-
Mitochondrial Function, Metabolic Regulation, and Human Disease Viewed through the Prism of Sirtuin 4 (SIRT4) Functions.J Proteome Res. 2019 May 3;18(5):1929-1938. doi: 10.1021/acs.jproteome.9b00086. Epub 2019 Apr 8. J Proteome Res. 2019. PMID: 30913880 Free PMC article. Review.
Cited by
-
Mechanism of Action of Ketogenic Diet Treatment: Impact of Decanoic Acid and Beta-Hydroxybutyrate on Sirtuins and Energy Metabolism in Hippocampal Murine Neurons.Nutrients. 2020 Aug 8;12(8):2379. doi: 10.3390/nu12082379. Nutrients. 2020. PMID: 32784510 Free PMC article.
-
Mitochondrial sirtuins in the heart.Heart Fail Rev. 2016 Sep;21(5):519-28. doi: 10.1007/s10741-016-9570-7. Heart Fail Rev. 2016. PMID: 27295248 Review.
-
Study of expression analysis of SIRT4 and the coordinate regulation of bovine adipocyte differentiation by SIRT4 and its transcription factors.Biosci Rep. 2018 Dec 14;38(6):BSR20181705. doi: 10.1042/BSR20181705. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30442871 Free PMC article.
-
Metabolic transitions regulate global protein fatty acylation.J Biol Chem. 2024 Jan;300(1):105563. doi: 10.1016/j.jbc.2023.105563. Epub 2023 Dec 13. J Biol Chem. 2024. PMID: 38101568 Free PMC article.
-
SIRT1 in the brain-connections with aging-associated disorders and lifespan.Front Cell Neurosci. 2015 Mar 9;9:64. doi: 10.3389/fncel.2015.00064. eCollection 2015. Front Cell Neurosci. 2015. PMID: 25805970 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
