A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation
- PMID: 24043761
- PMCID: PMC3782045
- DOI: 10.1084/jem.20130103
A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation
Abstract
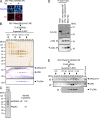
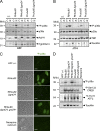
Toxoplasma gondii, the causative agent of toxoplasmosis, is an obligate intracellular protozoan parasite that resides inside a parasitophorous vacuole. During infection, Toxoplasma actively remodels the transcriptome of its hosting cells with profound and coupled impact on the host immune response. We report that Toxoplasma secretes GRA24, a novel dense granule protein which traffics from the vacuole to the host cell nucleus. Once released into the host cell, GRA24 has the unique ability to trigger prolonged autophosphorylation and nuclear translocation of the host cell p38α MAP kinase. This noncanonical kinetics of p38α activation correlates with the up-regulation of the transcription factors Egr-1 and c-Fos and the correlated synthesis of key proinflammatory cytokines, including interleukin-12 and the chemokine MCP-1, both known to control early parasite replication in vivo. Remarkably, the GRA24-p38α complex is defined by peculiar structural features and uncovers a new regulatory signaling path distinct from the MAPK signaling cascade and otherwise commonly activated by stress-related stimuli or various intracellular microbes.
Figures








References
-
- Bougdour A., Durandau E., Brenier-Pinchart M.P., Ortet P., Barakat M., Kieffer S., Curt-Varesano A., Curt-Bertini R.L., Bastien O., Coute Y., et al. 2013. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 13:489–500 10.1016/j.chom.2013.03.002 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

