Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes
- PMID: 24043818
- PMCID: PMC3791787
- DOI: 10.1073/pnas.1308049110
Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes
Abstract
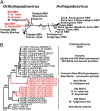
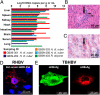
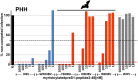
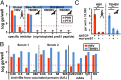
The hepatitis B virus (HBV), family Hepadnaviridae, is one of most relevant human pathogens. HBV origins are enigmatic, and no zoonotic reservoirs are known. Here, we screened 3,080 specimens from 54 bat species representing 11 bat families for hepadnaviral DNA. Ten specimens (0.3%) from Panama and Gabon yielded unique hepadnaviruses in coancestral relation to HBV. Full genome sequencing allowed classification as three putative orthohepadnavirus species based on genome lengths (3,149-3,377 nt), presence of middle HBV surface and X-protein genes, and sequence distance criteria. Hepatic tropism in bats was shown by quantitative PCR and in situ hybridization. Infected livers showed histopathologic changes compatible with hepatitis. Human hepatocytes transfected with all three bat viruses cross-reacted with sera against the HBV core protein, concordant with the phylogenetic relatedness of these hepadnaviruses and HBV. One virus from Uroderma bilobatum, the tent-making bat, cross-reacted with monoclonal antibodies against the HBV antigenicity determining S domain. Up to 18.4% of bat sera contained antibodies against bat hepadnaviruses. Infectious clones were generated to study all three viruses in detail. Hepatitis D virus particles pseudotyped with surface proteins of U. bilobatum HBV, but neither of the other two viruses could infect primary human and Tupaia belangeri hepatocytes. Hepatocyte infection occurred through the human HBV receptor sodium taurocholate cotransporting polypeptide but could not be neutralized by sera from vaccinated humans. Antihepadnaviral treatment using an approved reverse transcriptase inhibitor blocked replication of all bat hepadnaviruses. Our data suggest that bats may have been ancestral sources of primate hepadnaviruses. The observed zoonotic potential might affect concepts aimed at eradicating HBV.
Keywords: evolution; metagenomics; reverse genetics; virome; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30(12):2212–2219. - PubMed
-
- Chen DS. Toward elimination and eradication of hepatitis B. J Gastroenterol Hepatol. 2010;25(1):19–25. - PubMed
-
- Alter HJ. To have B or not to have B: Vaccine and the potential eradication of hepatitis B. J Hepatol. 2012;57(4):715–717. - PubMed
-
- Paraskevis D, et al. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57(3):908–916. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

